Drug repurposing by hetnet relationship prediction
A long time ago in a galaxy far, far away….
It is a dark time for drug discovery. The Empire spends over a billion dollars in R&D per new drug approval. The process takes decades, 9 out of 10 attempts fail, and the cost has been doubling every 9 years since 1970.
But, a small band of Rebel scientists pursue an alternative. Using public data and open source software, the Rebels are predicting new uses for existing drugs. Repurposing drugs avoids the main costs of drug development and is much faster since the drugs are already available and known to be safe.
The Rebels integrated data from every corner of the galaxy. Their hetnet contains 50 thousand nodes of 10 labels and 3 million relationships of 26 types. The Force allows a Data Jedi to predict which drugs treat which diseases. However to learn the Force—also known as a machine learning classifier—the Rebels need to summarize the network connectivity between each drug and disease. Join them in using neo4j to extract the features needed to learn the Force.
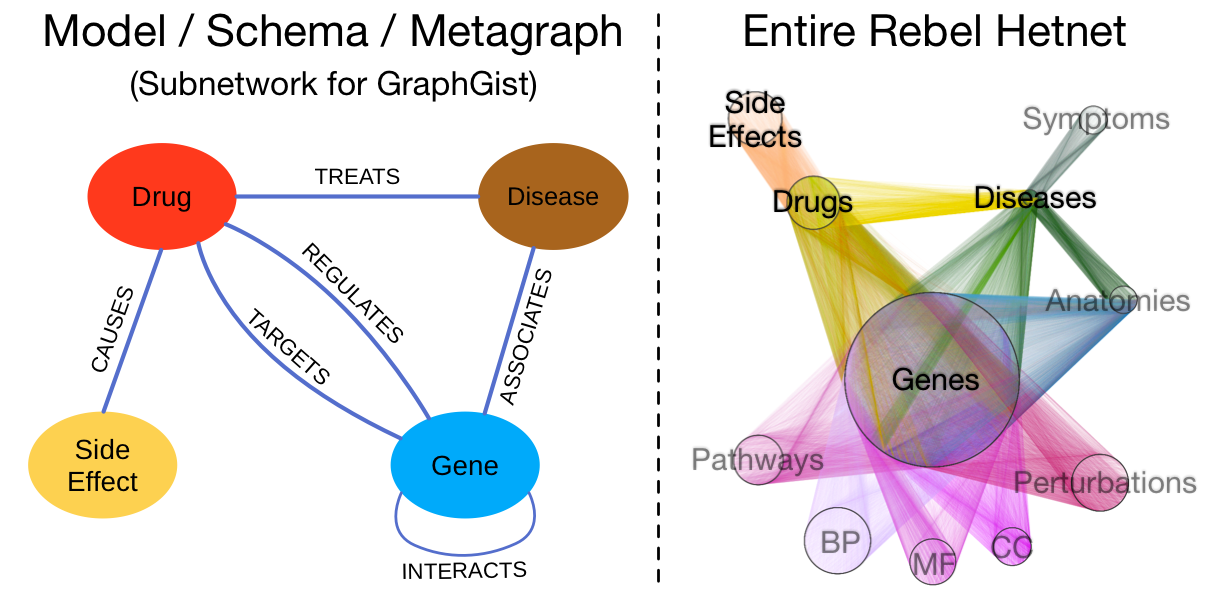
A subnetwork of the Rebel hetnet
Since the complete Rebel hetnet consists of 3 million relationships, it takes the entire Alliance Fleet to store. However, we’ve constructed a small illustrative subnetwork made to fit inside a single GraphGist starship. The left-hand image below shows the data model for the subnetwork, which contains four node labels and 6 relationship types. On the right, the entire Rebel hetnet is visualized from hyperspace: nodes are laid out orbitally by label and relationships are colored by type. The labels that are omitted in the subnetwork are in gray. We include this image to show the full progress of the Rebellion.
Cypher is the query language of the neo4j database. The following Cypher query creates the subnetwork for this GraphGist. It’s hidden by default, but you can click the expand arrows to see it.
The subnetwork used in this GraphGist is shown below. Use your lightsaber to reposition the nodes for a better view.
The example network contains 3 TREATS relationships. Between the 3 drugs and 2 diseases, there are six possible treatments (drug–disease pairs). The goal is to identify network patterns that distinguish the 3 present from 3 missing TREATS relationships.
Specifically, the Data Jedi Youngling searches for types of paths that occur more frequently between treatments than non-treatments. Here, we’ll investigate three path types (metapaths):
-
(:Drug)-[:TARGETS]-(:Gene)-[:ASSOCIATES]-(:Disease) -
(:Drug)-[:REGULATES]-(:Gene)-[:INTERACTS]-(:Gene)-[:ASSOCIATES]-(:Disease) -
(:Drug)-[:CAUSES]-(:SideEffect)-[:CAUSES]-(:Drug)-[:TREATS]-(:Disease)
Will these path types be sufficient to use the Force?
Drug targets and disease-associated genes
Both drugs and diseases relate with genes. Drugs target genes by binding to the encoded proteins. Diseases associate with genes when a gene plays a role in or determines susceptibility to a disease. One approach to drug discovery is identifying drugs which target genes associated with a disease. This concept is expressed by the (:Drug)-[:TARGETS]-(:Gene)-[:ASSOCIATES]-(:Disease) metapath. The following query counts the number of paths of this type for each drug–disease pair:
// Find all drug-disease pairs
MATCH (n0:Drug), (n2:Disease)
// Extract paths where the drug targets a gene associated with the disease
OPTIONAL MATCH paths = (n0:Drug)-[:TARGETS]-(n1:Gene)-[:ASSOCIATES]-(n2:Disease)
RETURN
// Retrieve the name of the drug and disease
n0.name AS drug,
n2.name AS disease,
// Retrieve whether the drug treats the disease
size((n0)-[:TREATS]-(n2)) AS treatment,
// Count the number of paths between the drug and disease
count(paths) AS path_count
// Sort the rows
ORDER BY path_count DESC, treatment DESCThe query finds one path and it’s between Clonidine and hypertension. Clonidine happens to treat hypertension suggesting that identifying drugs which target associated genes is a good repurposing strategy. However, the applicability of this approach is low: the other two known treatments have a path count of zero. Therefore, the Padawan must look to other path types with better coverage.
Gene regulation and interactions
Verifying drug targets requires time-consuming experiments that aren’t yet fully automatable. Therefore, this relationship type is highly incomplete—a common phenomenon in biological networks. However, recent high-throughput technologies have been able to more comprehensively relate drugs to genes. A recent project called LINCS profiled thousands of drugs and measured which genes change in abundance after cells are exposed to each drug. A drug is said to regulate a gene if the drug either increases or decreases the number of transcripts corresponding to that gene.
Another method for increasing the coverage of a path type is to increase its length. When proteins encoded by two genes form physical bonds inside a cell, the genes are said to interact. Genes tend to interact with other genes that perform similar functions, so adding an INTERACTS relationship to a metapath shifts the focus from a single gene to a neighborhood of functionally related genes.
Tying these sources together is the (:Drug)-[:REGULATES]-(:Gene)-[:INTERACTS]-(:Gene)-[:ASSOCIATES]-(:Disease) metatpath. Starting with a disease, the involved genes are detected by looking for genes that interact with associated genes. Then drugs are identified which regulate these genes. The goal is to find drugs which interfere with a gene neighborhood implicated in a disease.
// Find all drug-disease pairs
MATCH (n0:Drug), (n3:Disease)
// Extract paths following the specified metapath
OPTIONAL MATCH paths = (n0:Drug)-[:REGULATES]-(n1)-[:INTERACTS]-(n2)-[:ASSOCIATES]-(n3:Disease)
WITH
// reidentify the source and target nodes
n0 AS source,
n3 AS target,
paths,
// Extract the degrees along each path
[
size((n0)-[:REGULATES]-()),
size(()-[:REGULATES]-(n1)),
size((n1)-[:INTERACTS]-()),
size(()-[:INTERACTS]-(n2)),
size((n2)-[:ASSOCIATES]-()),
size(()-[:ASSOCIATES]-(n3))
] AS degrees
RETURN
// Retrieve the name of the drug and disease
source.name AS drug,
target.name AS disease,
// Retrieve whether the drug treats the disease
size((source)-[:TREATS]-(target)) AS treatment,
// Compute the path count
count(paths) AS path_count,
// Compute the degree-weighted path count with w = 0.5
sum(reduce(pdp = 1.0, d in degrees| pdp * d ^ -0.5)) AS DWPC
// Sort the rows
ORDER BY DWPC DESCWe now have two drug–disease pairs with at least one path. Since they’re both treatments, this feature appears predictive.
In the above query, we also calculate the degree-weighted path count (DWPC) for each drug–disease pair. The DWPC is a modification to the path count, which downweights paths through highly connected nodes. By rewarding highly specific relationships, which tend to be more informative, degree weighting can improve predictiveness. A single parameter, set here to 0.5, controls the strength of the weighting. For the best Jedi training, try learning the DWPC algorithm from its Cypher implementation. If that fails, see panel D of this diagram.
Side effects
FDA-approved drugs are required to list known side effects. Rebel researchers used text mining to catalog the side effects for all approved drugs, which we include in our hetnet. Side effects paint a high-level picture of a drug’s mechanism, regardless of whether the underlying molecular targets are known. One hypothesis is that drugs with similar side effects are likely to treat the same diseases. The (:Drug)-[:CAUSES]-(:SideEffect)-[:CAUSES]-(:Drug)-[:TREATS]-(:Disease) metapath looks for drugs that share side effects with a drug known to treat a disease.
Since approved drugs have abundant side effects, this feature is more complete than the previous two. All but one drug–disease pair has at least a single path. Several even have two paths. The top two DWPCs correspond to treatments, suggesting side effects can inform drug repurposing. The third ranked pair, Clonidine and glaucoma, is also a treatment although this knowledge wasn’t in our subnetwork. This illustrates the promise of our approach. Currently, many effective treatments are unknown, so the top ranking drug–disease pairs that are not current treatments are the ideal place to look for drug repurposing candidates.
Closing
We’ve computed features for three different path types in Cypher. In all three cases, paths were more prevalent between treatments than non-treatments. However, any individual path type was insufficient to separate all treatments from non-treatments. Thus the Jedi Knight uses the Force to combine information from many path types into a predictive classifier.
Weak alone are the features. Integrate and use the Force you must; the glue to bring diverse datasets together. Predict you will the probability that each drug treats each disease. But beware of the dark side. Relational databases and secrecy are the path to the dark side. Make open data and use neo4j you should.
If you’re interested in this project, visit the Rebel base to learn more.
© 2016, Daniel Himmelstein, released as CC-BY
Is this page helpful?