DeepWalk: Implementing Graph Embeddings in Neo4j

Artificial Intelligence Intern, Neo4j
11 min read

Almost a decade ago, Neo4j took off as a transactional graph database management platform. Just a year after its initial launch, a new, powerful query language called Cypher was introduced to allow expressive, efficient querying and other interactions with the graph data in Neo4j.
Today, Neo4j supports graph analytics including implementation of a wide variety of state-of-the-art graph algorithms with custom optimizations for Neo4j’s graph database model. It achieves unmatched performance in scalability, routinely running millions or even billions of nodes and edges.
The next natural step for the platform in 2019 is to venture into machine learning (ML) and deep learning.
The Evolution of AI
Artificial intelligence (AI) first launched when researchers came up with handcrafted features in an attempt to capture relevant attributes of various entities in the world.
Based on these entity models, sets of rules were manually written, aiming to encode how entities interact in the real world and predict how events may pan out in the future based on past and recent events.
The AI research community eventually realized that the world was too complex to be captured in terms of hand-coded attributes and interactions, as this approach was too slow and laborious to ever scale with respect to size and complexity.
This realization led to a major paradigm shift. Conventional AI was soon completely replaced by ML, in which entities and their attributes were hand coded, but rules governing their behaviors and interactions were automated.
This automation was largely based on statistical models, where only a general class of the model was specified, but its specific parameters and configurations were left for computers to learn from past data with various learning algorithms.
Today, even machine learning is a thing of the past. The dominant school of thought in artificial intelligence is deep learning. Deep learning automates both steps of the process: learning about both representation or entities, as well as the rules that govern their behaviors and interactions with each other.
This post will focus on the first step – the automated learning of representations (or embeddings). Specifically, we’ll look at a few different options available for implementing DeepWalk – a widely popular graph embedding technique – in Neo4j.
Graph Embeddings
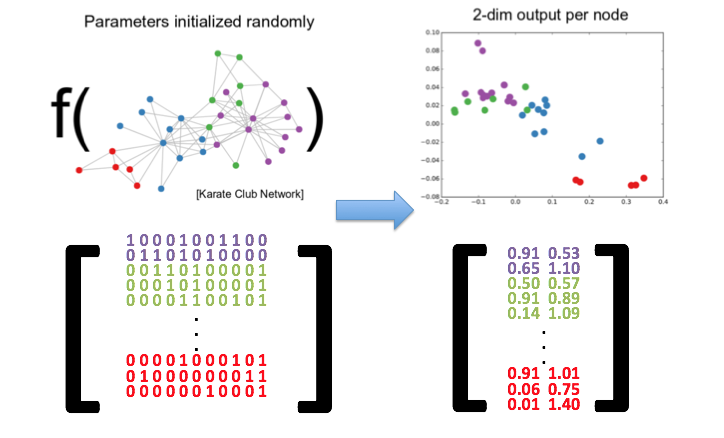
Embeddings transform nodes of a graph into a vector, or a set of vectors, thereby preserving topology, connectivity and the attributes of the graph’s nodes and edges. These vectors can then be used as features for a classifier to predict their labels, or for unsupervised clustering to identify communities among the nodes.
Example: Is Jon Snow really a Stark?
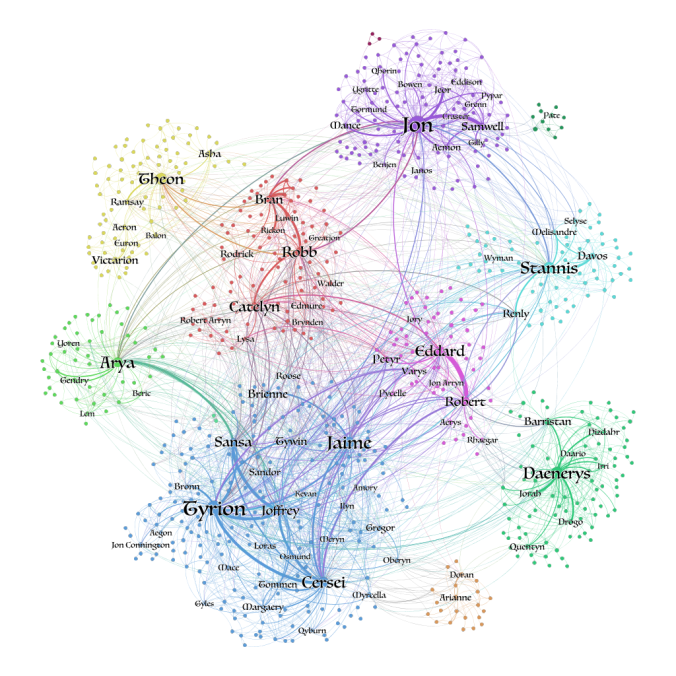
Let’s say we had a Game of Thrones (GoT) graph, where each node was one of the characters, and the edges between them encoded the characters’ interactions.
Now, if we were to try to predict the house label of the Jon Snow character, how would we go about it? Formally, this is a node classification problem, where we have to predict a node’s missing label or attribute.
If we were to produce an answer to this question using query-based feature engineering, we might try to retrieve the most common houses of Jon’s neighboring nodes and just take the most common one.
Similarly, if we were to use algorithms-based feature engineering to answer this question, we might run a community detection algorithm – such as Louvain – and associate the majority label of the node’s community with Jon Snow.
However, with graph embeddings, we would first learn embeddings to retrieve an embedded vector for each node in the graph, and then simply perform the following linear arithmetic: Jon - (Rob Stark + Ned Stark).
And that’s it! This roughly translates into the following query: “Rob is to Ned as Jon is to _______?”
Image from towardsdatascience.com
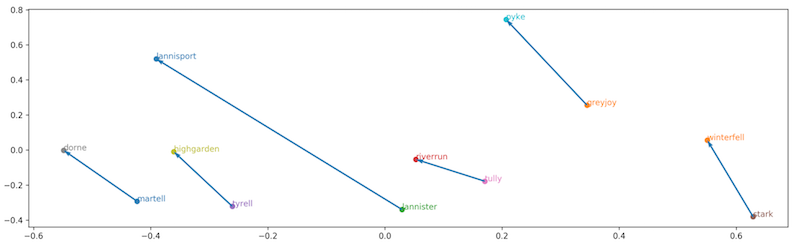
What is it about the embedded space that allows us to do this? Well, in the learning process of the embeddings, node are mapped onto the embedded space in a manner such that the different relationship between nodes are encoded in their relative positions.
Image from towardsdatascience.com
Attention to this phenomenon was first introduced by the seminal Word2Vec paper that kickstarted the rise of embeddings. In this paper, the author noted that in the embedded space, king - man + woman landed us closest to queen.
This is one of many examples that display how deep learning representations can be so powerful and encode semantic relations in a completely unsupervised manner.
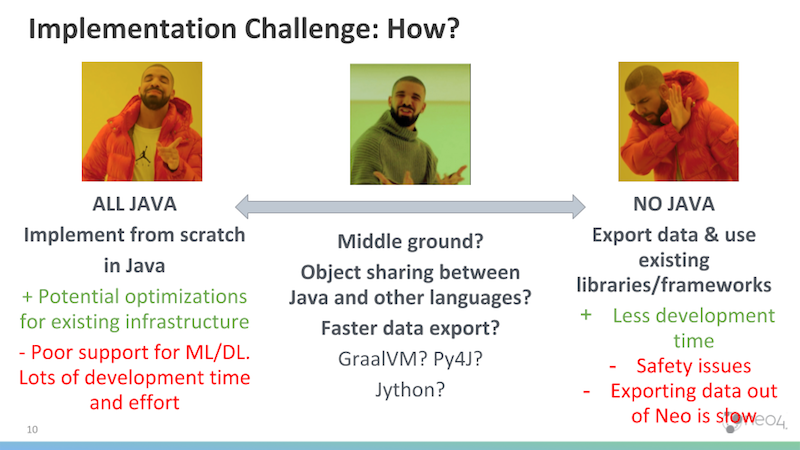
Up to this point, we’ve discussed what graph embeddings are and how they can be useful, but the question remains as to how we can implement them in Neo4j.
One of the major implementation challenges in this regard is that Neo4j – much like most enterprise level software in production – is written in Java, while most graph embeddings coming out of research labs remain in Python.
In the rest of this blog post, we explore the different options for implementing embeddings in Neo4j. We cover the entire spectrum, from Java to Python to hybrid approaches that allow object sharing between the two languages. Specifically, we focus on DeepWalk embeddings.
DL4J: All in Java
Deeplearning4j is an open-source, distributed deep-learning library in Java, and serves as Java’s equivalent to more popular deep learning frameworks in other languages such as Tensorflow and PyTorch, among others.
DL4J comes with many out-of-the-box deep learning implementations, including DeepWalk and node2vec, two of the most popular graph embeddings.
However, on deeper inspection of DL4J’s implementation of DeepWalk, we realized that some key components of DeepWalk were missing. The most important one was the complete absence of the “number of walks” hyperparameter, which controls the number of walks that are to be conducted starting from each node of the graph.
In its absence, the default implementation was only performing one random walk per node, which is not nearly enough. Resultantly, the learned embeddings were unable to attain a node classification accuracy score comparable to the numbers in the paper.
The code with the added “number of walks” hyperparameter is as follows:
@Override public List> getGraphWalkIterators(int numIterators){ List > list = new ArrayList<>(numIterators); int nVertices = graph.numVertices(); #New instance variable numIterators = this.numberOfWalks; #Random walks per node now equal to `numberOfWalks` for(int i=0; i < this.numberOfWalks; i++){ GraphWalkIterator iter = new RandomWalkIterator<>(this.graph, this.walkLength, this.rng.nextLong(), this.mode); list.add(iter); } return list; }
The complete implementation of DeepWalk using DL4J can be found on Github. This is implemented as a Neo4j plugin that can be downloaded in the Neo4j client and can be run as follows:
CALL embedding.dl4j.deepWalk();
This reads in the whole graph and writes back embeddings for each node as node properties, which can then be used for any downstream task. It is important to note that in our benchmarking experiments, we found DL4J implementations to be much slower than equivalent deep learning algorithms on other platforms.
Jython & JyNI
Jython is an implementation of Python that enables it to run on the Java virtual machine (JVM). This allows for the integration of Python code with Java libraries and other Java-based frameworks. It is important to note here that the default and most widely-used implementation of Python is in the C programming language and goes by Cython.
There are a number of key differences between Cython and Jython, perhaps the most significant being that Jython does not work with C extensions. Therefore, if a Python module utilizes C extensions, they do not work in Jython.
Likewise, Jython code runs seamlessly under CPython unless it contains Java integration. The lack of support for C extensions means Jython does not support key Python libraries for scientific computing and linear algebra, such as Numpy and Scipy.
To plug this gap, JyNI is a compatibility layer with the explicit goal of enabling Jython to make use of such native CPython extensions. In this way, scientific Python code can run on Jython.
However, JyNI, as of the time in which this post is published, also does not yet fully support Scipy and newer versions of Numpy, and there has been no activity on the Github repository for a year ¯_(ツ)_/¯.
Py4J: The Middle Ground
Py4J is a hybrid between using a glorified remote procedure call and the Java virtual machine to run a Python program.
The goal of Py4J – much like Jython – is to enable developers to program in Python and benefit from Python libraries, while also reusing Java libraries and frameworks.
However, as opposed to Jython, Py4J does not execute the Python code in the JVM, so it does not need to reimplement the Python language. In this way, developers can use all libraries supported by their Python interpreter, such as libraries written in Cython.
The Spark platform also uses Py4J for PySpark for object sharing between various Python and Java contexts.
Image from cwiki.apache.org
In terms of performance, Py4J has a bigger overhead than both the previous solutions because it relies on sockets. But, if performance is critical to your application, accessing Java objects from Python programs might not be the best idea.
Py4J is based on a server-client model that allows object sharing between the two. It basically allows users to set up a server in Java code and have the Python client read in objects in the JVM using local network sockets.
This object sharing can take place in both directions, as you can find the code for a working prototype for both the server and the client. In the prototype, a Python Py4J server for DeepWalk needs to run, and a Neo4j plugin in Java makes requests to it by passing the list of relationships in the graph and retrieving the node embeddings.
The following snippet is the Java code for the Neo4j plugin that serves as a Py4J client. It passes the graph as a list of relationships and retrieves embeddings from the DeepWalk Py4J server running in Python:
GatewayServer server = new GatewayServer();
server.start();
EmbeddingsInterface embeddings = (EmbeddingsInterface) server.getPythonServerEntryPoint(new Class[] { EmbeddingsInterface.class });
HashMap embs = embeddings.getEmbeddings(db.getAllRelationships());
for(Map.Entry m:embs.entrySet()){
Node n = db.getNodeById(Long.parseLong(m.getKey().toString()));
n.setProperty("emb", m.getValue());
}
Here’s the code for the Py4J server for DeepWalk in Python that accepts a list object containing relationships from Java clients:
from py4j.java_gateway import JavaGateway, CallbackServerParameters, GatewayParameters,GatewayClient
def load_py4jclient(relationships, undirected=True):
rels = list(relationships)
G = Graph()
for rel in rels:
x, y = rel.split("_")
G[x].append(y)
if undirected:
G[y].append(x)
G.make_consistent()
return G
gateway = JavaGateway(gateway_parameters=GatewayParameters(),
callback_server_parameters=CallbackServerParameters(),
python_server_entry_point=embs)
GraalVM: The Middle Group
GraalVM is a universal virtual machine for running applications written in JavaScript, Python, Ruby, R. It also runs applications in JVM-based languages like Java, Scala, Groovy, Kotlin, Clojure, as well as LLVM-based languages such as C and C++. GraalVM removes the isolation between programming languages and enables interoperability in a shared runtime.
Graal’s support for Python, however, is questionable at this point. They don’t shy away from constantly reminding you of this through their documentation, especially when one runs:
graalpython *.py and the following message is printed:
Please note: This Python implementation is in the very early stages, and can run little more than basic benchmarks at this point.
No surprises here, but DeepWalk cannot run on GraalVM Python, which depends on relatively sophisticated libraries such as Gensim, which is in turn dependent on Cython, Scipy, Numpy and other libraries.
Calling Python (or Any Bash Call) from Java
With Jython, we tried communication between Java and Python code using sockets. Here, we go old school and simply use the file system. The code is fairly simple though a little hacky.
A Java Neo4j plugin exports the graph as a comma-separated list of edges onto the local file system. External code is called via bash shell, passing the exported comma-separated values (CSV) as a parameter like so:
Process process = Runtime.getRuntime().exec(new String[]{
condaPath+"/envs/deepwalk/bin/deepwalk",
"--format", "edgelist",
"--input", path+"/data/tmp.csv",
"--output", path+"/data/tmp.embeddings"
});
Finally, the embeddings CSV file is read back from the file system and written to the graph as node properties:
@Procedure(name="example.deepWalk", mode=Mode.WRITE)
@Description("Calls deepwalk from bash and writes back embeddings as node properties")
public void deepWalk() throws IOException, FileNotFoundException, InterruptedException, KernelException{
String condaPath = System.getenv("CONDA");
String dir = System.getenv("NEO4J_HOME");
//write out graph as edgelist
exportData(dir);
//call bash command
learnEmbeddings(dir, condaPath);
//read back embeddings csv as node properties
loadEmbeddings(dir);
}
The complete code can be found on Github. Naturally, this approach does not scale well; the cost of exporting the whole graph as a CSV and reading back an embeddings CSV can be prohibitively expensive for large graphs.
In addition, it also raises concerns of data security , specifically regarding the process of writing out graphs as plain text.
Python Drivers: All in Python
This approach lies at the other extreme end of the spectrum from where we started. It does not involve any Java, and all the code is in Python.
The original implementation of DeepWalk comes with support for reading data in different formats, though all expected on the file system. The supported formats include edgelists, adjacency matrix and others. These are specified using the --format flag.
Here, we use Neo4j drivers to add provision for an additional input format: --format neo.
When this flag is turned on, data is read from a running Neo4j instance (assuming default IP, port and credentials) rather than from a file on the file system. After the learning phase is complete, the embeddings are written back as node properties to the Neo4j database, using the drivers.
Complete code can be found here. The code involving drivers is given below:
import py2neo
from py2neo.database import Graph as NeoGraph
def load_neo(undirected=True):
G = Graph()
graph = NeoGraph()
rels = list(graph.match())
for rel in rels:
x, y = int(rel.start_node.identity), int(rel.end_node.identity)
G[x].append(y)
if undirected:
G[y].append(x)
G.make_consistent()
return G
def write_to_neo(keyedVectorEmbeddings):
graph = NeoGraph()
nodes = list(graph.nodes.match())
subgraph = None
for node in nodes:
emb = list(keyedVectorEmbeddings[str(node.identity)])
node['embedding'] = str(emb)
subgraph = subgraph | node if subgraph is not None else node
graph.push(subgraph)
Benchmarking Experiments
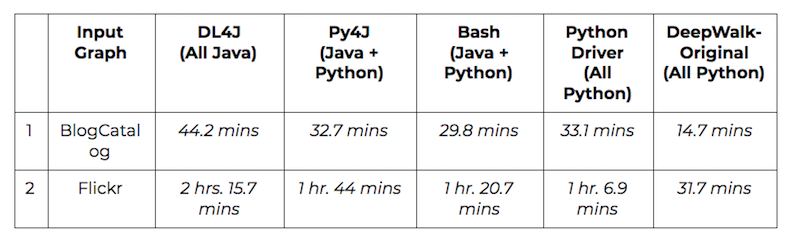
All the various options for introducing embeddings to Neo4j – DeepWalk, specifically – were benchmarked against two graph datasets, commonly cited in academic papers on graph embeddings: BlogCatalog (10K nodes, 330K edges) and Flickr (80K nodes, 5.8M edges, 195 node labels). The benchmarking experiments were run on 2.6 GHz Intel Core i5 with 8 GB 1600 MHz DDR3.
The following table gives CPU run times for each approach on the two datasets:
Conclusion
In this post, we used DeepWalk and Python as case studies for how existing open-source implementations of different graph embeddings can be brought into Neo4j.
Given the slow times, it may appear that re-implementing these implementations – often in C or Python – again in Java, using DL4J for instance, is not a good idea. This is probably the main takeaway from this post.
Also, note that these prototypes/proof-of-concepts are nowhere near optimal implementation, nor were the benchmarking experiments conducted in the most sterile testing environments. This post and the POCs are more springboards than the final word.
Show off your graph database skills to the community and employers with the official Neo4j Certification. Click below to get started and you could be done in less than an hour.