My Neo4j Summer Road Trip to the World of Healthcare [Part 4]

Editor-in-Chief, Neo4j
11 min read
Part 4: Create Relationships with FuzzyWuzzy
Welcome back to my adventure to the world of healthcare! In the past three blog posts, I have discussed the data model of the healthcare graph, loading XML data into Neo4j and cleaning CSV data in the command-line interface. In this post, I will demonstrate how to link data from multiple data sources, especially when there is a lack of foreign IDs to identify records.
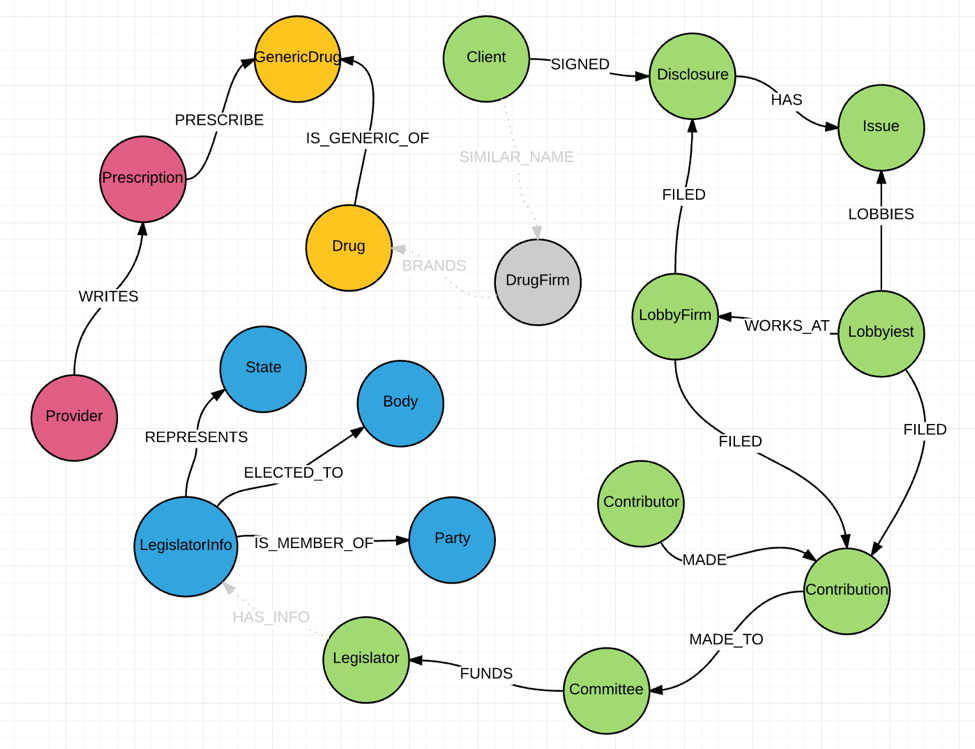
The healthcare graph consists of four groups of data, with each group of nodes generated from different data sources. Let’s start off by looking at the four groups of nodes and how I created relationships within the group.
1. Lobby Disclosure Nodes Group
This group is extracted from public lobbying disclosures, which includes these nodes from our healthcare data model: (:Disclosure), (:Client), (:Issue), (:Lobbyist), (:LobbyFirm), (:Contribution), (:Contributor), (:Committee) and (:Legislator). While extracting the nodes from the original XML files, nodes IDs are generated internally by Neo4j. As a result, I could use the internal node IDs to create relationships to connect these nodes. I have documented this process in my second blog post.
2. Legislator Nodes Group
This group includes nodes (:LegislatorInfo), (:State), (:Body) and (:Party), which are extracted from a single CSV file that can be downloaded here. Relationships are created through a single Cypher
statement during the ETL process. All the ETL code can be found at GitHub here.
3. Provider Prescription Nodes Group
Nodes (:Prescription) and (:Provider) are generated from CMS data sources. I stored National Provider Identifiers as a property {NPI} for both nodes and used it to connect (:Provider) with (:Prescription).
4. Drug Nodes Group
Nodes (:GenericDrug) and (:Drug) are extracted from FDA data sources. Both nodes along with (:Prescription) have property {GenericName}. (:GenericDrug) is created as an intermediate node to present each unique {GenericName} value in (:Drug).
RxNorm is a national medical library that provides a RESTful API which allows me to link clinical drug vocabularies to normalized names such as Rxcui, a unique ID for drugs. I use the batch mode to send {GenericName} of (:Prescription) and (:GenericDrug) to get Rxcui and connect these two nodes on the drug ID Rxcui.
Nodes can be simply connected together if they are extracted within a single file or from the same source. Using standardized unique IDs is even more convenient to join nodes together. However, pulling data from a variety of sources and having limited access to data are often the cases that data journalists or data engineers need to tackle.
The issue I am trying to demonstrate in this post is how to create relationships to connect different groups of data. Specifically, how to link (:Client) and (:DrugFirm), (:DrugFirm) and (:Drug), as well as (:Legislator) and (:LegislatorInfo).
Graph databases emphasize representation of relationships among data points. Without the relationships, I won’t be able to create the whole path of the healthcare graph and would lose the ability to track information following by these paths. The idea of connecting the nodes, such as (:Drug) and (:DrugFirm) is that if the drug is branded by a drug firm, a relation should exist to connect these two nodes together.
Now let’s take a look of the properties of these two nodes and we may find some useful information.
MATCH (d:Drug), (df:DrugFirm) RETURN d as Drug, df as DrugFirm LIMIT 25
(:Drug) has a property {labelerName} and (:DrugFirm) contains {firmName}. The logic of connecting a drug firm with a drug is the {firmName} can be recognized as identical or similar to the {labelerName}. This may sound like an easy task for human beings to do, but automating this process can be a little tricky.
Luckily I have heard of FuzzyWuzzy, a practical Python package, which does string matching and returns the matching rate. I decided to give it a try to match the {firmName} with {labelerName}.
1. Array Structure
The first step before comparing two arrays of strings is to structure the array to make it easy to work with:
#======= RETURN Drug object: list of dics, key: labelerName, id ======#
q1 = '''
MATCH (d: Drug)
RETURN id(d), d.labelerName
'''
drug_obj = g.run(q1)
drugs_lst = []
for object in drug_obj:
drug_dic = {}
drug_dic['id'] = object['id(d)']
drug_dic['labelerName'] = object['d.labelerName']
drugs_lst.append(drug_dic)
#======= RETURN DrugFirm object: list of dics, key: firmName, id ======#
q2 = '''
MATCH (df:DrugFirm)
RETURN id(df), df.firmName'''
df_obj = g.run(q2)
df_lst = []
for object in df_obj:
df_dic = {}
df_dic['id'] = object['id(df)']
df_dic['firmName'] = object['df.firmName']
df_lst.append(df_dic)
Here I returned an array of dictionaries for both (:Drug) and (:DrugFirm). Each dictionary represents an object, and each object has two keys: Node internal ID and {labelerName} / {firmName}. The node internal ID is used later on to fetch the nodes that we want, which we will talk about shortly. The {labelerName} and {firmName} are the strings that we will compare. Now let’s print out the data structure:
DrugFirm:
[{'id': 23049075, 'firmName': 'Teva Branded Pharmaceutical Products R&D, Inc.'},
{'id': 23049076, 'firmName': "George's Family Farms, LLC"},
{'id': 23049077, 'firmName': 'Baxter Healthcare S.A.'},
{'id': 23049078, 'firmName': 'Tokuyama Corporation'},
{'id': 23049079, 'firmName': 'Alps Pharmaceutical Ind. Co., Ltd.'}]
Drug:
[{'id': 22941414, 'labelerName': 'Eli Lilly and Company'},
{'id': 22941415, 'labelerName': 'Eli Lilly and Company'},
{'id': 22941416, 'labelerName': 'Eli Lilly and Company'},
{'id': 22941417, 'labelerName': 'Eli Lilly and Company'},
{'id': 22941418, 'labelerName': 'Eli Lilly and Company'}]
2. String Preprocessing
As we could see, some company names are in uppercase and some are in lowercase. Some of the names contain non-alphanumeric characters. There are also many duplicates in the array, which will slow down the string matching process.
To improve the matching rate, I passed the arrays to a series of string processing functions to clean up the strings.
#lower case: convert all to lower case lc_ln = lower_case(drugs_lst, 'labelerName') lc_fn = lower_case(df_lst, 'firmName') #remove_marks: remove non-alphanumeric characters rm_ln = rm_mark(lc_ln, 'labelerName') rm_fn = rm_mark(lc_fn, 'firmName') #Chop_end: remove ‘s’ at the end of a string ce_ln = chop_end(rm_ln, 'labelerName', 's') ce_fn = chop_end(rm_fn, 'firmName', 's') #sort_strings: sort words in a string sort_ln = sort_strings(ce_ln,'labelerName') sort_fn = sort_strings(ce_fn, 'firmName') #uniq strings: de-duplicate: collect nodes ID in to a list for each unique string uq_ln = uniq_elem(sort_ln, 'labelerName') uq_fn = uniq_elem(sort_fn, 'firmName')
All the functions in the script can be found here. After processing the strings, I de-duplicated the objects in {labelerName} and {firmName}, the number of objects are decreased from 106683 to 5928 for {labelerName} and 10205 to 7040 for {firmName}.
Now let’s print out the uq_ln and uq_fn to understand the structure before we move on.
uq_ln:
defaultdict(, {'akorn llc stride': [22965573, 22965574, 22965575, 22965576], 'brand inc silver star': [23040080, 23040084, 23040091, 23040097, 23040151, 23040169, 23040171, 23040172, 23040174], 'co ltd osung': [23007551, 23007552, 23007553], 'beauticontrol': [23013024], 'biological glaxosmithkline sa': [23013557, 23013558, 23013559, 23013560, 23013561, 23013562, 23013563, 23013564, 23013565, 23013566, 23013567, 23013568, 23013569, 23013570, 23013571],…
uq_fn:
defaultdict(, {'biological glaxosmithkline sa': [23053065], 'bioservice capua spa': [23052723], 'american homepatient': [23057681, 23057683, 23057686, 23057687, 23057688, 23057689, 23057690, 23057691, 23057692, 23057693, 23057694, 23057695, 23057696, 23057697, 23057698], 'healthcare limited novarti private': [23056487],…
I have two defaultdicts to work with. The keys in each array are the strings to be compared, the values are node IDs representing the nodes that contains the same strings (i.e., the key). Now I can call FuzzyWuzzy to get string matching rates.
3. FuzzyWuzzy String Matching
for k1 in uq_ln:
labeler_name = k1
nodeId_drug = uq_ln[k1]
for k2 in uq_fn:
company_name = k2
nodeId_df = uq_fn[k2]
r1 = fuzz.partial_ratio(labeler_name, company_name)
r2 = fuzz.ratio(labeler_name, company_name)
if r1 > 85:
print('r1:',r1, 'r2:',r2, 'ln:',k1, 'fn:',k2)
if r2 > 85:
print('r1:',r1, 'r2:',r2, 'ln:',k1, 'fn:',k2)
I started off by choosing 85 to be the cut off for both partial_ratio and ratio just to see the results of the string matching. I generalized the result into three cases:
Case 1: Both partial_ratio(r1) and ratio(r2) are equal to 100
The two strings are identical to each other, for example:
r1: 100 r2: 100 ln: abilene inc oxygen fn: abilene inc oxygen r1: 100 r2: 100 ln: abilene inc oxygen fn: abilene inc oxygen
Case 2: Only r1 is 100
One of the strings is a substring of the other one, but needs to exclude false positives. Here is the example:
r1: 100 r2: 65 ln: gavi llc pharmaceutical fn: llc pharmac
From my observation on the example, fn is the substring of ln and r1 is equal to 100, supporting my observation. However, the r2 is relatively low. By judging from the two strings, I cannot infer they represent the same company. Thus we need to do some further modification either on the cut off rate or the strings to exclude the false positives.
Case 3: Both r1 and r2 are >85
The two strings are similar, but need to exclude false positives:
r1: 98 r2: 96 ln: barr inc laboratorie fn: arg inc laboratorie r1: 89 r2: 94 ln: company perrigo fn: company l perrigo
Both r1 and r2 are greater than 85 in the two examples above. However, I identify the first line being a false positive, which needs to be excluded. Whereas in the second line, the two strings may represent the same company even though both r1 and r2 are lower than the first line’s. Again, we need some further modifications to improve the accuracy of the string matching.
4. Modification on String Matching
Let’s take a look at the strings in case 2 and case 3 again:
“barr inc laboratorie fn” vs “arg inc laboratorie” -> false positive
“company perrigo” vs “company l perrigo” -> may represent the same company
I want to do some changes on the strings so my program will only return the third line and ignore the first two lines. I realize most of the company names are constructed by two components: a unique component (in bold) that differentiates the company from other companies, and a common component (in regular text) that represents the type of organization.
If I remove the common component from the string, whatever is left should be the unique component which is much more precise and easier for a computer to decide whether the two strings are similar or not.
For example, if remove all the common components in all the three lines, the strings will look like this:
“barr” vs “arg” -> false positive
“perrigo” vs “l perrigo” -> may represent the same company
Now the computer can pick up the third line (r1 improved from 89 to 100) where “perrigo” is a substring of “I perrigo”, and it can ignore the first two false positive cases.
I also noticed for most of the cases when r1 = 100 and r1–r2 > 30, it’s hard to say if the two strings represent the same company, such as in this example:
r1: 100 r2: 24 ln: brand inc tween fn: br r1: 100 r2: 43 ln: bio co general ltd fn: bio c r1: 100 r2: 47 ln: dava inc pharmaceutical fn: ava inc r1: 100 r2: 59 ln: bio cosmetic fn: bio c r1: 100 r2: 27 ln: chartwell governmental llc rx specialty fn: al llc r1: 100 r2: 69 ln: canton laboratorie fn: canton laboratorie limited private r1: 100 r2: 48 ln: beach inc productsd fn: ch inc r1: 100 r2: 63 ln: genentech inc fn: ch inc r1: 100 r2: 41 ln: dental llc scott supply fn: al llc
As a result, I decided to exclude these cases by controlling the differences between the cut off values within the range of 30.
5. The Final Solution
# #======= Create relation :BRANDS (String Fuzzying Matching) ======#
q3 = '''
MATCH (d:Drug) where id(d) in {drug_id} and d.tradeName is not NULL
MATCH (df:DrugFirm) where id(df) in {drug_firm_id}
MERGE (df)-[r:BRANDS]->(d)
ON CREATE SET r.ratio = {r2}, r.partial_ratio = {r1}'''
num = 0 # Number of rel that are created
for k1 in uq_ln:
labeler_name = k1
nodeId_drug = uq_ln[k1]
for k2 in uq_fn:
company_name = k2
nodeId_df = uq_fn[k2]
r1 = fuzz.partial_ratio(labeler_name, company_name)
r2 = fuzz.ratio(labeler_name, company_name)
if r1 == 100 and (r1 - r2) <= 30:
g.run(q3, drug_id = nodeId_drug, drug_firm_id = nodeId_df, r1 = r1, r2 = r2)
num += 1
print("CREATE relation :BRANDS number:", num)
elif (100 > r1 >= 95 and r2 >= 85) or (95 > r1 >= 85 and r2 >= 90): ### miss spell or miss a word r1 and r2 > 95
md_r1 = fuzz.partial_ratio(string_filter(labeler_name, nostring), string_filter(company_name, nostring))
md_r2 = fuzz.ratio(string_filter(labeler_name, nostring), string_filter(company_name, nostring))
if md_r1 >= 95 and md_r2 >= 95:
g.run(q3, drug_id=nodeId_drug, drug_firm_id=nodeId_df, r1=md_r1, r2=md_r2)
num += 1
print("CREATE relation :BRANDS rel number:", num)
I decided to create the relationship [:BRANDS] for nodes (:Drug) and (:DrugFirm) if r1 is 100 and r1–r2 is less than 30. When r1 and r2 are both above 85, I decided to filter out some common words in the strings, such as inc, co, ltd, llc, corporation, pharmaceutical, laboratory, company, product, pharma, etc. and then recalculate r1 and r2 on the modified strings.
It’s also helpful to store the value of r1 and r2 as properties for [:BRANDS], so when I am querying information between drug firms and drugs, I am also able to trace the confidence level of the answer. The Cypher below returns the {firmName} and {labelerName} based on the ascending order of r1:
MATCH (df:DrugFirm)-[r:BRANDS]->(d:Drug) RETURN df.firmName, d.labelerName, r.partial_ratio as r1, r.ratio as r2 order by r1 ASC limit 10
Now we have successfully created the relationship [:BRANDS] for (:DrugFirm) and (:Drug), and the matching results seem pretty trustworthy. I also used the same method to create relationships for (:Client) and (:DrugFirm), as well as (:Legislator) and (:LegislatorInfo).
Lastly, let’s find out which drug firm brand drug has the generic component morphine sulfate:
MATCH (df:DrugFirm)-[r:BRANDS]->(d:Drug{genericName:'Morphine Sulfate'}) RETURN df,d
(There are 193 matched results and I randomly choose 3 companies to display the results.)
Conclusion
I hope you enjoy reading through this blog post. If you are facing similar issues such as having difficulties connecting to your graph, I hope this post is helpful to you.
At the same time, I am eager to hear your thoughts or ideas on how to solve the problem. Don’t be hesitate to send me a message on Twitter, LinkedIn or email if you have any questions about my project.
With the summer passing by, it is getting close to the end of my fun summer road trip through the world of healthcare. I will be very excited to show you some the most interesting discoveries I have made in my next blog post. If you want to know more about graph technology in the healthcare industry, don’t miss out on my last blog post. See you soon!
Ready to dig in and get started with graph technology? Click below to download this free ebook, Learning Neo4j and catch up to speed with the world’s leading graph database.
Catch up with the rest of the Neo4j in healthcare series: