From Text to a Knowledge Graph: The Information Extraction Pipeline

Graph ML and GenAI Research, Neo4j
10 min read

Editor’s note: This presentation is given by Tomaz Bratanic at NODES 2021.
As organizations build knowledge graphs to find answers to their most pressing problems, one of the challenges they face is that much of the information they would like to incorporate in their knowledge graphs exists in unstructured text data, such as news articles, emails and scientific journal entries.
Building an information extraction pipeline allows a developer to take these texts as inputs, process them with NLP (Natural Language Processing) techniques, and use the resulting structures to populate or enrich their knowledge graph. In this blog, I will explain how to build an information extraction pipeline to transform unstructured text inputs into a useful knowledge graph.
How It Works
To begin, I input text to the pipeline. This can be text stored in PDFs or scraped from web pages, new slides, text files, or even emails if you’re a journalist conducting an investigation. If you’re in a business environment, you could also input messages from your messaging platform into the information extraction pipeline.
From there, you would get a knowledge graph.

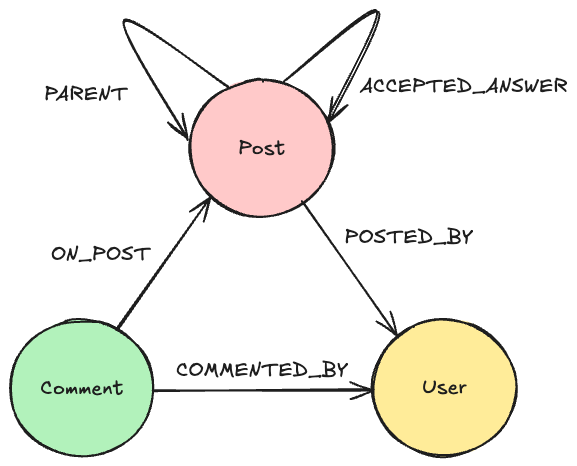
A graph has nodes that represent levels, entities, or concepts. There are many types of nodes, such as person, course, or city, and there are relationships between those nodes, such as located in, lives in, or works with. If you have a single relationship type, you are not yet dealing with a knowledge graph until you have more relationship types.
4-Step Pipeline

The input is text, and the output is a knowledge graph. To get there, I’ve implemented an information extraction pipeline. This example was a hobby project.
My information extraction pipeline consists of four steps. The first three steps are NLP techniques, and then we store the results of the NLP techniques into a graph database. As you’ve probably gathered, I will be focusing more on how to construct a knowledge graph with the help of NLP techniques, rather than how to analyze a knowledge graph.
Constructing a knowledge graph is important because there is a wealth of information generated every day. Businesses have emails or PDFs with large amounts of text generated in addition to relevant texts available on the internet, including photos and scrape pages. In the future, information extraction pipelines will become more and more relevant.
Step 1: Coreference Resolution
The first step is the coreference resolution, which is an NLP language technique that finds all expressions that refer to the same entity in a text. It finds all the pronouns and then replaces those with their reference entities. Pronouns can be personal, location, or possessive. Here’s a simple example.
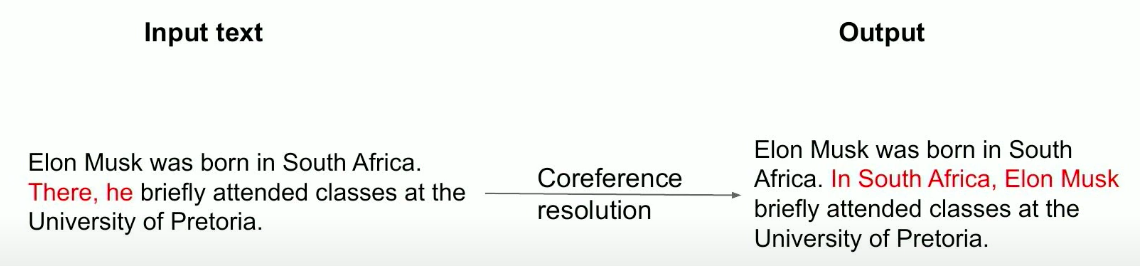
We can input: Elon Musk was born in South Africa. There, he briefly attended classes at the University of Pretoria. You can see that the first sentence doesn’t have any pronouns, but the second sentence has two pronouns, one locational and one personal. The ideal result of the coreference resolution model would be an output that replaces There with South Africa and replaces he with Elon Musk. We end up with: In South Africa, Elon Musk briefly attended classes at the University of Pretoria.
This might seem really basic, but real-world examples of coreference resolution could get quite complicated. Plus, it is a very important step in the information extraction pipeline.
Step 2: Named Entity Recognition
Once we’ve dealt with the coreference resolution, we move on to named entity recognition, which is a task for recognizing all the mentioned entities in the text. Those entities could be of various types; for example, we could have a person, location, or geopolitical entity.

We also have organizational entities, which will find the entities that the NLP model is trained on. The training data really depends on the domain or the intended obligations that you’re using your information extraction pipeline for. There are a couple of limitations for named entity recognition. I will just point out the most obvious with the example I’ve shown using Elon Musk in three different sentences.
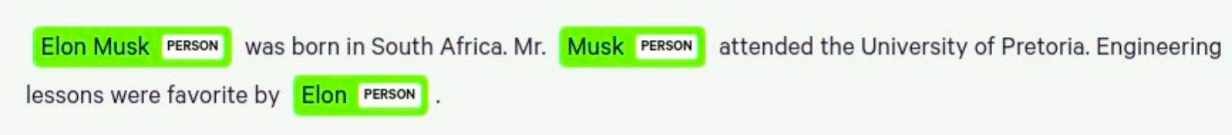
The first sentence uses Elon Musk’s full name, and the second time, he’s mentioned as Mr. Musk. In the third sentence, he’s mentioned as Elon. In an output knowledge graph, we ideally want a single real-world entity to be represented with a single node. We really don’t want to have three different nodes representing the same entity as you would have here. Remember, the named entity recognition order depends on the domain of the intended application. Most of today’s open-sourced models are based on different types of texts. For example, if you wanted to analyze biomedical text and extract biomedical concepts, like genes, drugs, or diseases, then you would need to use a different NLP model that was trained to extract biomedical concepts.
No model is 100% accurate. Even when I was preparing this example, if in the third sentence, I started the sentence with Elon to say Elon’s favorite lessons were engineering, then Elon was not recognized as a person in the spacing model.
Entity Disambiguation and Entity Linking
How could we solve the entity disambiguation so that a single real-world entity would be represented as a single node in a knowledge graph? The answer is entity linking. To go from name entity recognition to name entity linking means that we aggregate the name entity recognition to first recognize all the entities, and then map those entities to a target knowledge base. Very frequently, that target knowledge base is Wikipedia. In essence, you recognize the name entity linking model as entities in the text, and then link them to a Wikipedia ID.
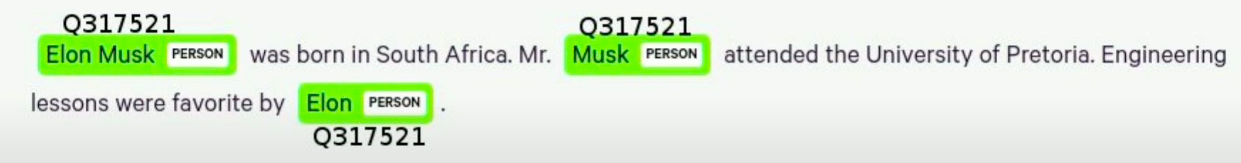
Here you can see that I’ve added Wikipedia ID for all the entities. Ideally, the result would be that even though we have three different titles of texts for Elon Musk, they would all link to the same entity on Wikipedia. We could use that Wikipedia ID as identities for nodes in our knowledge graph, and we would end up with a single node in our knowledge graph.
This process is so common that linking entities to Wikipedia is actually called “wikification”. The plus side is we don’t have to worry about which text record is correct because here we have three different text records that refer to the same entity. We could just use the Wikipedia ID and then use the different texts and names, and link them all back to the entity name on Wikipedia.
Whilst we have Wikipedia IDs for the entities, we can also use Wikipedia to enrich our knowledge graph. You could combine the information that you get from the information extraction pipeline with online information available on Wikipedia, and use that to create a more full knowledge graph, starting with NLP techniques. Some of you might say that Wikipedia is not the most accurate source, but Google uses Wikipedia extensively. So I say, if it’s good enough for Google, it’s good enough for me.
Co-Occurrence Graphs
Co-occurrence networks for graphs are basically when you’re inferring relationships between a pair of entities based on their presence within a specified unit of text.

For example, we could say that if two entities co-occur or are present in the same sentence, that they are somehow related. We don’t really know how, but we know that they are somehow related because they appeared in the same sentence. Here you can also see that the coreference resolution is quite important; if we split the specified unit of text into a sentence and don’t use coreference resolution here in the second sentence, we’ll have a single entity with two pronouns. That would lead to our information extraction not being full, but we can avoid this by using coreference resolution.
Step 3: Relationship Extraction
What if we want to extract semantic relationships from text and don’t just want a generic relationship between them, where they all just co-occur? We want to define a semantic type to the relationship between a pair of entities.

The relationships are usually categorized for the readability of your narrative. There are two different approaches to categorizing relationships between pairs of entities. One approach is the rule based relationship extraction, where you use grammatical dependencies of the texts to extract relationships. The second approach is to train an NLP model to extract relationships between a pair of entities. First, we’ll take a look at rule-based relationship extraction.
Rule-Based Relationship Extraction
I’ve used spacing to visualize the grammatical dependencies of the sentence, Elon Musk was born in South Africa. RDF (Resource Description Framework) graphs use a data selection tree post, where they have a subject, verb, predicate, and object. The predicate is the relationship between the two entities.
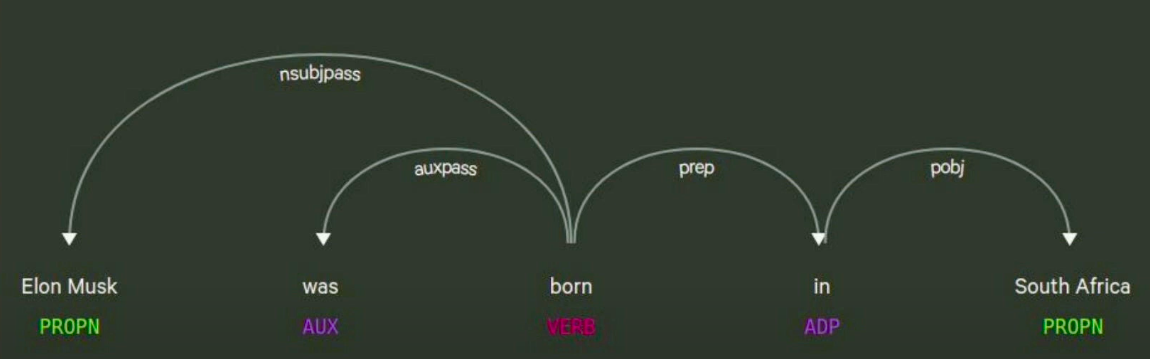
As you can see in the image above, we have the verb in the middle, and the subject is Elon Musk. The object is South Africa. We can infer that Elon Musk was born in South Africa instead of just having a co-occurrence generic relationship between them. There are some pros and cons to this. You’re not really doing any semantic categorization of your relationships because you’re just taking the verb, which has the time component, meaning it could be present, future, or past tense. This may or may not be a good idea to have in a knowledge graph . There could be synonyms of verbs that have different relationship types with similar, identical semantic values.
My suggestion would be that if you use the rule-based relation extraction, then you also should do some post-processing of the verbs and try to fill them into some semantic categories. This way you don’t end up with so many relationship types. For example, you could use synonyms to drop some verbs into the same category.
NLP Model Based Relationship Extraction
The other approach that applies to relation extraction is using an NLP model again. You have to train your NLP model to infer the relations between pairs of entities, and the types of relationships that I would infer depend on the training data that is used to train the model. For example, if you’re analyzing news articles, you want to have a different model than if you are analyzing biomedical articles. TACRED is a dataset made available by Stanford, and then there is the Wiki80 dataset, which we’ll now take a closer look at.
Training Your NLP Model Using Wiki80
The Wiki80 training dataset contains 80 different relationship packs. The same semantic categories of relationships used on Wikipedia are also used in the training data. For me, it’s really interesting to see that if you use Wikipedia as the target database for entity linking, you’re using the Wiki80 training dataset. You can further train the relationships that fall to the Wikipedia ontology between pairs of entities. If you do this, you’re actually constructing a long mini Wikipedia-like knowledge graph. I find that quite awesome!
Real World Example: Drug Repurposing
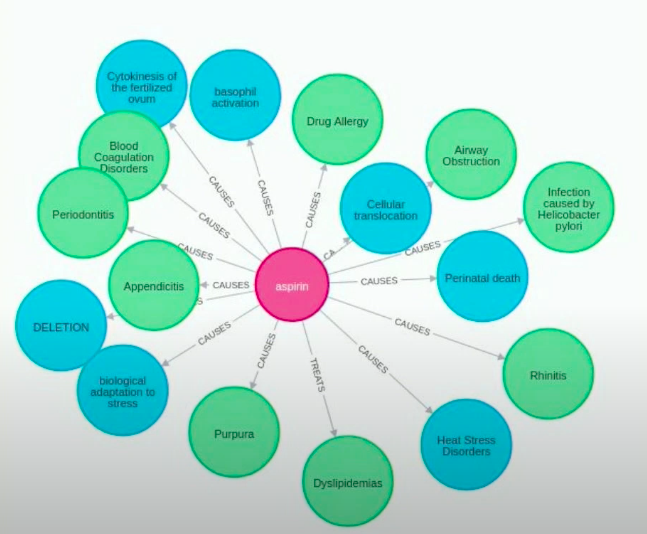
Now we’ll look at a real-world example of an information extraction pipeline for drug repurposing. You need a knowledge graph with medical concepts, so you can have many types of nodes. These node types could be genes, drugs, diseases, or other types of nodes. There are many types of relationships between these medical concepts, as shown above: TREATS, CAUSES, PREDISPOSES, and others.
Then you can make link predictions for the TREATS relationship, which would predict new uses for a drug. This is drug repurposing. You use text input into the information extraction pipeline, such as medical journals, including the PubMed dataset, or medical papers, and then perform a named entity linking. Like Wikipedia, you could use online medical databases, including ENSEMBL, to map genes to the corresponding IDs in the target database. There is also CHEBI for chemical substances. Basically, you do the name entity linking and extract your biomedical concepts. To extract relationships between those entities is challenging since there are no existing training datasets available, but there is a semantic MEDLINE Database where a group of scientists and doctors have a base relation extraction for data available on their website. You could also use that to try extracting relations with your own NLP model.
Step 4: Knowledge Graph
Now that you have biomedical concepts as entities and the relationships between them, you create a knowledge graph and try to predict new use cases for existing drugs.

Here, I’ve created a simple example where we have drugs and diseases. The real-world example is of course much more complicated because you could also have genes, meaning a headache could be associated with the same genes for migraines, and then there are relationships between genes. It quickly gets complicated, but I’ll just give you a simple idea for drug repurposing. When you try to predict new links, what you do is length reduction, or what is called a knowledge graph completion, since it’s quite different if this new predicted relationship is TREATS or CAUSES.
Conclusion
There you have it! You’ve used the information extraction pipeline to construct a knowledge graph. Now our information extraction pipeline expands the scope of data we can import into a knowledge graph.
From just being able to use structured data, we can now also use unstructured text data! This greatly expands the power, scope, and usefulness of our knowledge graphs. I hope you, too, after reading this blog, can find ways to incorporate text data into your knowledge graphs!
Thinking about building a knowledge graph?
Download The Developer’s Guide: How to Build a Knowledge Graph for a step-by-step walkthrough of everything you need to know to start building with confidence.