Leading Pharmaceutical Company Cuts Drug Discovery Timelines in Half with Neo4j-Powered Knowledge Platform on AWS
A global biopharmaceutical company transforms data into knowledge and improves target accuracy 50x with a Neo4j graph intelligence platform.
50%
faster clinical trial site selection
4 Hours vs. Years
to identify potential treatments for rare diseases
50x
more accurate drug target identification with proprietary ranking algorithm

In four hours, scientists at a leading pharmaceutical company identified two potential treatments for a rare endocrine disorder — a disease with no approved therapies. Their secret: a Neo4j-powered graph intelligence platform that makes drug discovery 50 times more accurate than random chance.
Behind every successful drug are thousands of medicines that never made it to market. Developing a single drug takes 15 years and $3.5 billion — yet only 6.2% of new drugs entering Phase I trials ever reach their intended users. For the 30 million Americans living with rare diseases, many spending decades without treatment options, every day counts.
Drug repurposing is one tool to address this challenge. Repurposing allows researchers to use decades of existing scientific and clinical knowledge to find new solutions for patients faster than starting from scratch. Yet even drug repurposing faces the same challenge that slows new drug development: knowledge fragmented across disconnected data systems, preventing breakthrough ‘aha!’ moments.
In 2019, the company decided to cut drug development timelines in half by rethinking how pharmaceutical research works from first principles. Their knowledge platform now unifies over 200 data sources in a Neo4j graph database that contains more than 100 million nodes and 1 billion relationships. Scientists now identify drug targets 50 times more accurately than random chance. They discover treatments in hours instead of years. They predict safety issues before testing begins.
According to company leadership, the platform “is an amazing database that integrates all the internal data from clinical [programs], notebooks from scientists, preclinical experiments, and also everything externally, and brings them together using machine learning and artificial intelligence to create actionable knowledge.”
The Hidden Crisis of Fragmented Knowledge
“We had a fundamental data fragmentation problem,” the company explains. “A neurology team and an oncology team could be investigating the exact same protein but from different angles, and their findings might never intersect because of structural barriers.”
Insights that could accelerate treatments for Parkinson’s, lupus, and cancer were trapped in isolated databases. Scientists spent weeks manually searching through publications looking for crucial connections. Leadership recognized that traditional approaches wouldn’t solve the fundamental problem: they needed to stop collecting data and start connecting knowledge.
“Let’s stop thinking about being data-driven, and instead think about being knowledge-driven,” company researchers noted when presenting the platform at a 2023 industry conference. “There’s a fundamental difference there.”
Knowledge preserves the important context clues that enable AI systems to make leaps in reasoning. The company recognized that NoSQL solutions alone couldn’t handle the interconnected nature of biomedical knowledge at the scale required for real-time discovery applications. While other databases excel at handling large volumes of data, they struggle with the multi-hop queries and relationship traversals essential for pharmaceutical research. The company needed a platform that could mirror how the real world operates.
Neo4j’s graph intelligence platform gave them this capability. Neo4j enables the company to model biomedical knowledge as it actually exists: a web of interconnected relationships between genes, diseases, compounds, clinical trials, and patient outcomes.
Teaching Machines to Think Like Scientists
Building the platform required five years of intensive work. Hundreds of scientists validated each knowledge assertion, ensuring accuracy across disciplines. The platform integrates 450 terabytes of medical knowledge from over 200 sources including clinical trial results, genomic databases, scientific publications, internal experimental data, and real-world patient outcomes.
Instead of storing simple facts like “gene A causes disease B,” the system captures context, for example: “gene A might cause disease B according to this study published in Science in 2023, based on specific experimental results, with a confidence score of 0.7.”
Neo4j’s Graph Data Science (GDS) library also serves as the platform’s analytical engine, providing over 65 ready-to-use algorithms that run directly on the connected data. The platform uses PageRank analysis to identify genes most strongly associated with specific diseases — the same algorithm that powers Google search.
“PageRank provides a probability that a random web surfer would come across your page,” the team explains. “We apply this same concept to find the genes most connected to a given health condition through multiple pathways in our knowledge graph.”
Drug Discovery
The platform’s proprietary ranking analysis has revolutionized how the company identifies drug targets. The algorithm analyzes the network of relationships connecting genes to diseases, weighing connections based on strength scores derived from scientific evidence. Researchers can steer the analysis using different weighting techniques, emphasizing clinical relevance for established therapies or novel relevance for cutting-edge research.
For complex conditions like lupus, the system shows an enrichment factor of 50 — meaning the algorithm identifies relevant genes 50 times more accurately than random selection.
As the research team explains, “If you had to pick genes associated with a given health condition randomly, you’d hit a percentage or two. Using Neo4j’s PageRank algorithm, we can identify genes that are 50 times more likely to be relevant than random picking.”
Drug Repurposing
In a striking demonstration of the platform’s speed, scientists identified potential treatments for a rare endocrine disorder — a deadly disease with no approved therapies — discovering two existing market drugs in just four hours using the platform’s integrated knowledge approach.
The platform also applies Neo4j’s graph algorithms to computational toxicology, using PageRank and link prediction to infer relationships between drug candidates and potential adverse events. This enables prediction of safety liabilities computationally before animal or human testing, allowing scientists to prioritize safer compounds earlier in development.
Process Automation
Machine learning algorithms analyzing the knowledge graph have improved clinical trial site selection by over 50%, increasing the number of sites meeting enrollment and startup goals while reducing selection timelines to 10-12 weeks. This improvement directly impacts patients by accelerating the clinical trial process and ensuring studies can be completed more quickly and efficiently.
According to the development team, “We’re using the platform at all stages of R&D development. It’s really the focal point of our digital ecosystem, and it’s increasingly being used by leadership to make decisions.”
Designed to Scale on AWS
The platform runs on AWS infrastructure designed for both massive scale and real-time responsiveness. Neo4j serves as the intelligent relationship layer, connecting rather than replacing existing systems. This architecture lets the company preserve investments in current infrastructure while adding graph intelligence capabilities.
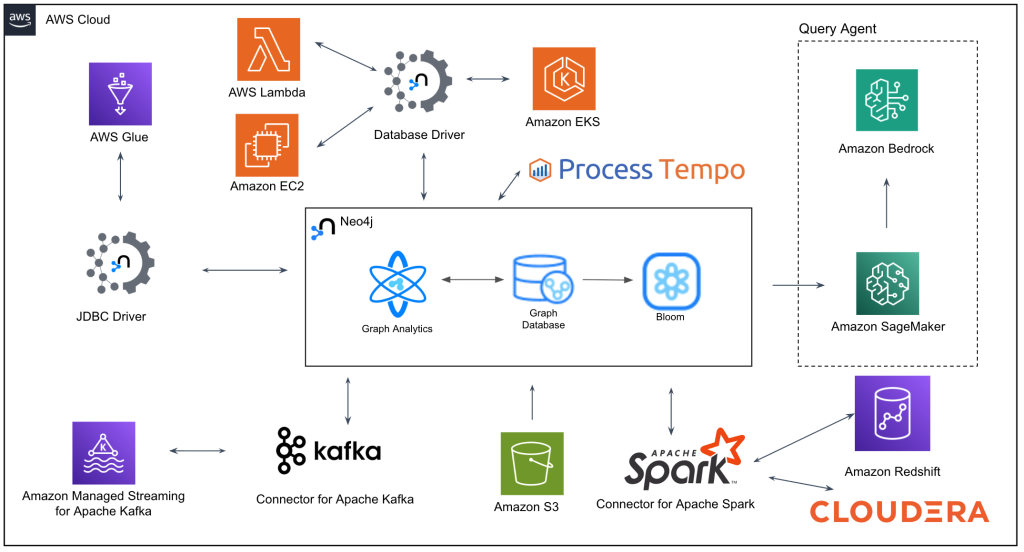
Above: The company’s AWS infrastructure
The platform processes hundreds of queries per minute, with most completing in under 500 milliseconds. Scientists get answers at the speed of thought, enabling exploratory analysis that would be impossible at such a scale with a NoSQL database. Real-time data streams from ongoing experiments continuously enrich the knowledge graph, ensuring researchers always work with current information.
Integration with Amazon Bedrock and SageMaker brings large language models into the workflow. Scientists can query the platform using natural language, with AI translating questions into graph traversals and presenting results with clear, actionable insights.
When a scientist asks a question in natural language, such as “What genetic markers are associated with acute myeloid leukemia?”, an AI agent first queries the knowledge graph to retrieve a set of verified facts and source documents related to the query. By forcing the LLM to generate its answer based on this specific, curated context from the graph, the system dramatically reduces the risk of AI hallucinations and ensures all answers are traceable and verifiable.
Third-party data management tools provide stakeholders with a simple, code-free interface to the graph environment. The platform team has developed several dashboards for tracking data lineage, data freshness, impact analysis, and to help stakeholders understand and explore the metadata. External systems can access this same information through standard APIs. The company is now working to deliver enhanced graph data visualization and exploration features for the next version of the solution.
Next Steps: Toward AI-Assisted Research
Every capability the platform adds translates directly to patient impact. Faster target identification means treatments reach trials sooner. Better safety prediction reduces failures that delay approvals. Improved trial selection ensures studies complete faster with better data. And the company’s vision extends far beyond current capabilities.
“The aim here is ambitious,” the team notes. “Accelerate drug development timelines by 40-50% and transform how science is done.”
The next phase involves AI agents that traverse relevant paths in the knowledge graph to retrieve facts and source documents — then generate concise, multi-level summaries. This creates a virtuous cycle where new insights continuously enrich the knowledge graph, making it progressively more powerful. The knowledge graph acts as a factual grounding layer, reducing the risk of AI hallucinations while ensuring traceability back to original sources.
The platform represents a new model for pharmaceutical discovery in a world where 95% of rare diseases lack approved treatments.
The company intends for the platform to generate its own research ideas, and eventually papers to support them. The system will read thousands of scientific papers, spot patterns in how diseases develop, and suggest new experiments that human scientists might never have considered. Instead of researchers spending months connecting dots between different studies, the platform could propose: “Based on 500 papers about protein interactions, this heart medication might also treat Alzheimer’s disease.”