Using Embeddings to Represent String Edit Distance in Neo4j

Senior Data Scientist at Neo4j
6 min read
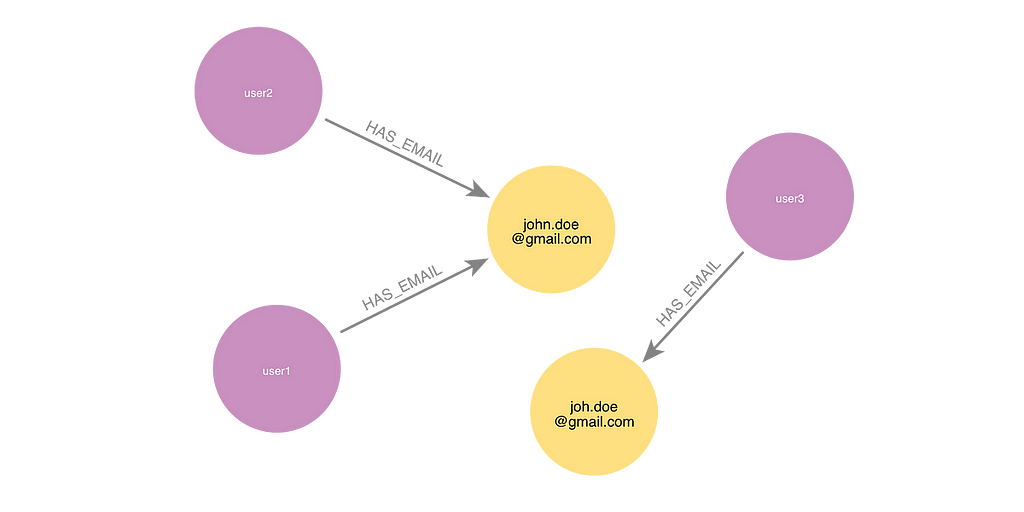
Graph databases like Neo4j are great for solving entity resolution problems. For example, a graph shows where two user profiles share a common identifier, like an email address. However, it can be challenging for a graph to identify cases where two identifiers are similar but with a slight spelling difference that might result from a data entry error.
Neo4j’s Awesome Procedures On Cypher (APOC) library has several text functions for computing string edit distance, including Levenshtein, Jaro-Winkler, and Sorensen-Dice. This works well for comparing individual pairs of strings, but would take a long time for a brute-force approach to apply these functions to every possible pair of strings in a large dataset.
I came across Convolutional Embedding for Edit Distance by Xinyan Dai, Xiao Yan, Kaiwen Zhou, Yuxuan Wang, Han Yang, and James Cheng. The authors used a convolutional neural network (CNN) to create an embedding vector that represents the spelling of a string. The CNN is trained to put strings that differ by a small edit distance closer together in the vector space than strings that differ by a large edit distance. Instead of needing to check all possible pairs of strings to find misspellings, we can use K-nearest neighbors to identify the most likely string pairs that should be compared.
I cloned the code from the GitHub repo that the authors provided. I added some Jupyter notebooks to execute the code and work with the resulting data in Neo4j. Check out my fork of the repo and the code for this blog.
Generate Synthetic Email Data
I used the Faker Python library to generate a list of 150,000 synthetic email addresses. After I removed duplicates, I found that I had 141,286 unique emails to work with. The code for generating the emails is in the graph_analysis/Generate_data.ipynb notebook.
Generate Embeddings and Send to Neo4j
Next, I ran the authors’ embedding code. The code splits the example data into train, query, and base segments and generates embeddings for each string. I made one change to the trainer.py file to make the default embedding architecture the two-layer CNN. The code I used to kick off the embedding is in the graph_analysis/Send_to_Neo4j.ipynb notebook.
python main.py --dataset emails --nt 1000 --nq 1000 --epochs 20 --save-split --save-embed --save-model
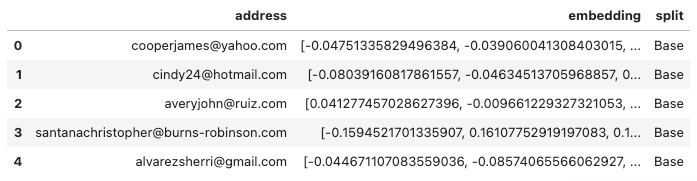
Generating the embeddings took about 93 seconds. Here’s an example of the results.
I created a node key constraint in Neo4j to make sure that each Email node in Neo4j would be required to have a unique address value.
CREATE CONSTRAINT email_node_key IF NOT EXISTS FOR (e:Email) REQUIRE e.address IS NODE KEY
Then I used a Cypher query to send the emails with their embeddings to Neo4j. It took about 67 seconds to load the data.
UNWIND $data AS row
CALL {
WITH row
MERGE (e:Email {address:row['address']})
SET e.split = row['split']
WITH row, e
CALL db.create.setNodeVectorProperty(e, 'editEmbedding', row['embedding'])
} IN CONCURRENT TRANSACTIONS OF 10000 rows
Use K-nearest Neighbors to Find Most Similar Embeddings
Neo4j’s Graph Data Science offering includes K-nearest neighbors. For each Email node in the graph, the algorithm will try to find the k other nodes with the most similar embeddings based on cosine similarity. The code to execute this part of the process is in the notebook graph_analysis/Check_knn.ipynb.
First, I used the Neo4j GDS Python client to create an in-memory projection of the Email nodes. Because there are no relationships in the database yet, I used a wildcard (“*”) for the relationship projection.
g_email, result = gds.graph.project(
"emails",
{"Email": {"properties": "editEmbedding"}},
"*")
I ran KNN to find the 10 other nodes with the most similar embeddings for each Email node. I compared the cosine similarity between the embeddings returned by KNN with the Levenshtein distance between the addresses returned by apoc.text.levenshteinDistance() from Neo4j’s APOC library.
top_10_df = gds.run_cypher("""
CALL gds.knn.stream("emails", {nodeProperties:"editEmbedding", topk:10})
YIELD node1, node2, similarity
WITH gds.util.asNode(node1) AS s,
gds.util.asNode(node2) AS t,
similarity
RETURN round(similarity, 2) AS similarity,
apoc.text.levenshteinDistance(s.address, t.address)
AS levenshteinDistance,
count(*) AS pairs
""")
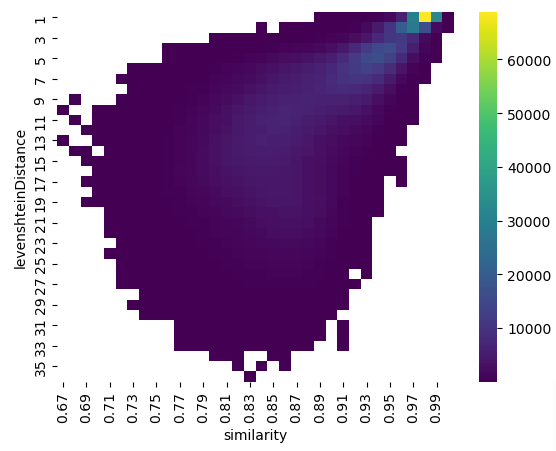
I created a heat map of the results. I could see that there was a relationship between embedding cosine similarity and string Levenshtein distance. None of the string pairs with a Levenshtein distance of two or less had an embedding similarity of greater than 0.84.
In my initial exploration, I looked at the top 10 most similar embeddings for each node, but in a large email dataset, an email might have more than 10 other emails within a small edit distance. I ran KNN again, this time with the top K parameter set to 80 and a similarityCutoff of 0.84. Instead of streaming the results, I wrote new relationships to Neo4j called HAS_SIMLAR_EMBEDDING. It took 270 seconds to calculate these results and write them to Neo4j. All together, creating the embeddings, sending them to Neo4j, creating the graph projection, and running KNN in write mode took 431 seconds, a little over seven minutes. Here is the Cypher query that I ran:
gds.run_cypher("""
CALL {
CALL gds.knn.stream("emails", {nodeProperties:"editEmbedding", topk:80, similarityCutoff:0.84})
YIELD node1, node2, similarity
WITH gds.util.asNode(node1) AS s,
gds.util.asNode(node2) AS t,
similarity
WITH s, t, similarity,
apoc.text.levenshteinDistance(s.address, t.address) AS levenshteinDistance
WHERE levenshteinDistance <= 2
MERGE (s)-[r:HAS_SIMILAR_EMBEDDING]->(t)
ON CREATE SET r.similarity = similarity,
r.levenshteinDistance = levenshteinDistance
} IN TRANSACTIONS OF 10000 ROWS""")
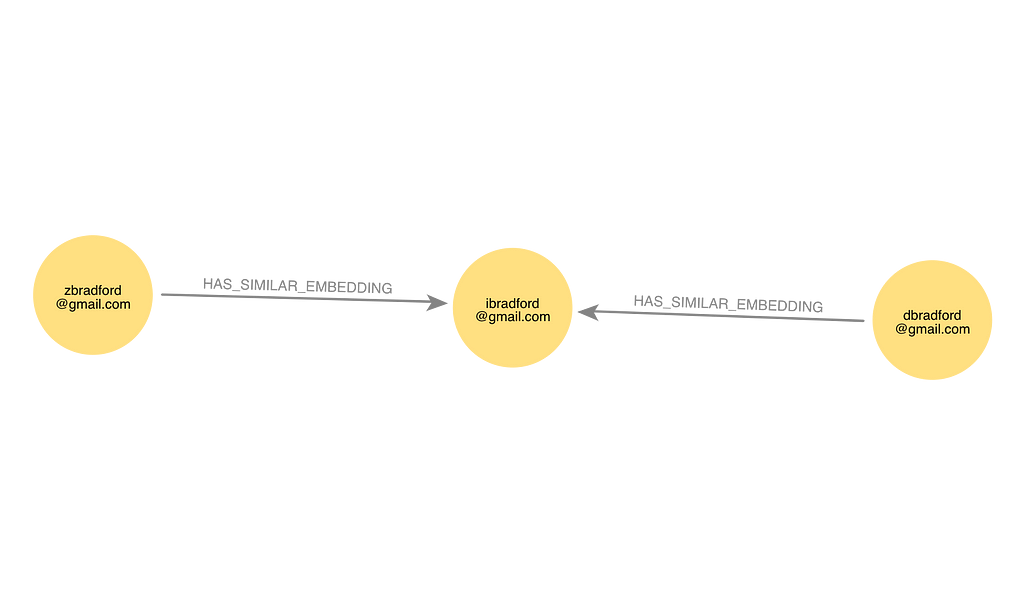
Here is an example of the output in Neo4j showing an Email node linked to two other nodes by HAS_SIMILAR_EMBEDDING relationships.
Compare KNN Results With Record Linkage Results
I wanted to compare those results with what I would get from using Python’s record linkage library. The code for this part of the process is in the graph_analysis/Try_record_linkage.ipynb notebook. Doing a comparison of every possible pair of nodes using a full index would be too memory-intensive for my environment. I loaded the emails into a Pandas dataframe, and I set up blocking columns.
First, I tried requiring that all potential matches have the first letter of the email in common. Running the record linkage indexer to create those candidate pairs caused my environment to run out of memory.
To cut down the number of potential matches, I required that candidate email pairs must have either the same first two characters and the same top-level domain or the same characters from the third character up until the top-level domain. I knew this would miss some pairs with edits in the first two characters or in the top-level domain, but hopefully, those types of mistakes would be relatively rare in real data. This made 64,816,224 possible pairs to be examined. Creating the index and running the comparison in record linkage took 609 seconds. This was 29 percent slower than the CNN/Neo4j approach.
I loaded the pairs from record linkage to Neo4j to see how closely the pairs from the embedding comparison matched the pairs from record linkage. I found that 248,790 pairs (92.8 percent of total) were found by both processes; 15,237 pairs (5.7 percent of total) were found by the embedding comparison but missed by record linkage; 4,106 pairs (1.5 percent of total) were found by record linkage but missed by embedding comparison.
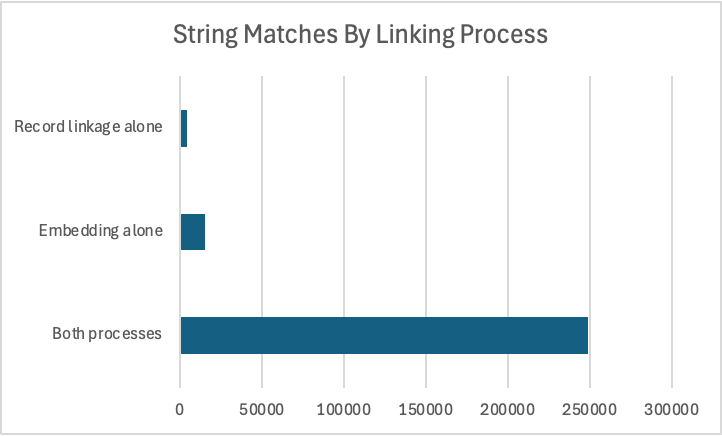
When embedding similarity found pairs that record linkage missed, I saw that it was because there was an edit within the first two characters of the string — amills@yahoo.com and zmiles@yahoo.com, for example. We would need more compute resources to try a different blocking scheme to avoid this problem in record linkage.
When record linkage found pairs that embedding comparison missed, it was because the CNN embedding placed addresses with a Levenshtein distance greater than 2 closer to the target address than the email with a Levenshtein distance of 2 or less. I might explore a more complex CNN architecture than the two-layer CNN to avoid this problem.
Overall, I was impressed with the results of using a CNN to identify pairs of strings with low edit distance. I think it has potential for inclusion in graph-based entity resolution pipelines.
Using Embeddings to Represent String Edit Distance in Neo4j was originally published in Neo4j Developer Blog on Medium, where people are continuing the conversation by highlighting and responding to this story.