“Science has explored the microcosmos and the macrocosmos; we have a good sense of the lay of the land. The great unexplored frontier is complexity.” – Heinz Pagels
Anyone working in the life sciences is well aware of the scale and complexity of the data generated by their work. This has long been the case and a well covered topic in both of our previous workshops. In an increasingly digitised world, finding meaning in ever growing, disparate datasets is fast moving beyond the capacity of a single human mind. The challenge is exacerbated by an explosion of openly available information like scientific publications, annotations, ontologies and APIs that may have a direct bearing on particular areas of interest.
Our third Neo4j Health Care & Life Sciences Workshop has been set up to showcase practical solutions to common problems as well as helping to incubate collaboration, innovation and good practice. Graph databases are powerful tools that are inherently capable of managing vast quantities of data points and the web of relationships between them. As people start turning to tools like Neo4j for answers there are inevitably more questions: data modeling, performance, resilience, interoperability. These are the kinds of questions we want to help you answer.
View the decks from the presentations of the workshop.

|

|
-
04:10:03

Neo4j Health Care & Life Sciences Workshop Day 1
Watch Video -
03:53:19
Neo4j Health Care & Life Sciences Workshop Day 2
Watch Video -
26:01
Neo4j Health Care & Life Sciences Workshop Day 1 – Opening
Watch Video -
24:05
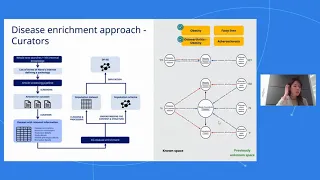
Graph data science and knowledge graphs for target discovery for cardiometabolic diseases
Watch Video -
20:56
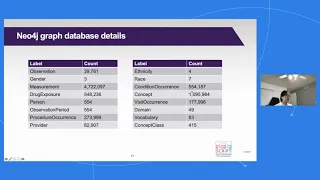
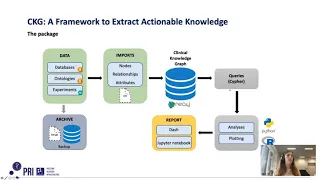
Mapping the OMOP Common Data Model to Neo4j for Pneumonia Therapy Response: SCRIPT Case Study
Watch Video -
25:24
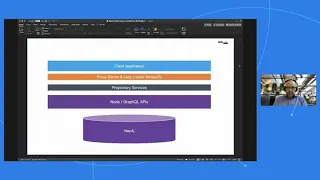
Building complex Web Applications for Health Care using Neo4j
Watch Video -
29:03
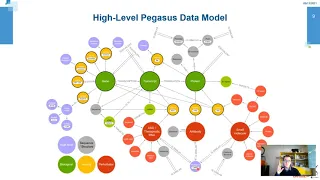
Pegasus: a knowledge graph to support drug discovery
Watch Video -
14:34
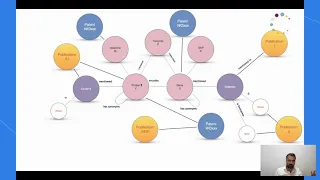
HealthECCO: Bringing the world’s health knowledge to research and medical decision makers
Watch Video -
15:55
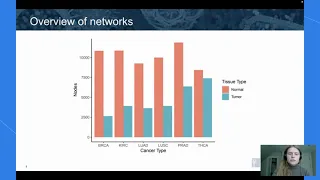
Analyzing Perturbed Co-Expression Networks in Cancer Using a Graph Database
Watch Video -
26:57
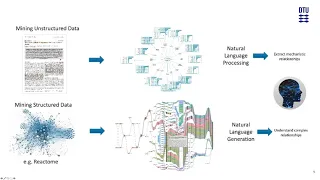
Lifelike – a graph-powered knowledge reconstruction and mining platform to accelerate research
Watch Video -
19:50
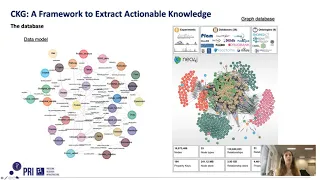
Graph-centric framework for translational clinical proteomics
Watch Video -
18:08
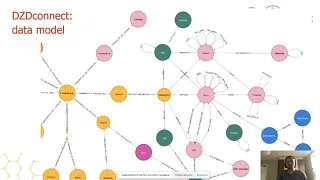
From Queries to Algorithms to Advanced ML: 3 Pharmaceutical Graph Use Cases
Watch Video -
22:43
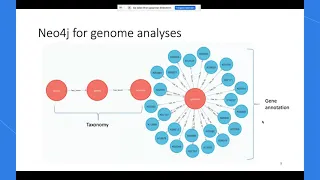
Neo4j for Bacterial Genomes
Watch Video -
25:51
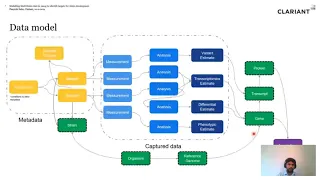
Modelling Multi Omics data in Neo4j to identify targets for strain development
Watch Video -
22:38
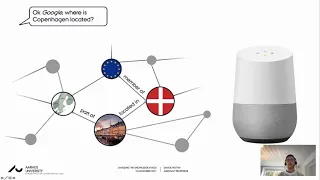
Unveiling the knowledge in knowledge graphs
Watch Video -
16:03

Evaluating Drug Safety Data Using Graph Databases
Watch Video -
38:23

Clinical Knowledge Graph with Spark NLP and Neo4j
Watch Video -
18:05

Neo4j Health Care & Life Sciences Workshop Closing Panel Discussion
Watch Video