Announcing: Graph-Native Machine Learning in Neo4j!
5 min read

We’re delighted to announce you can now take advantage of graph-native machine learning (ML) inside of Neo4j! We’ve just released a preview of Neo4j’s Graph Data Science™ Library version 1.4, which includes graph embeddings and an ML model catalog.
Together, these enable you to create representations of your graph and make graph predictions – all within Neo4j.
Until now, few companies outside of leading Big Tech have had the resources and ability to take advantage of advanced graph-based ML techniques. Neo4j for Graph Data Science™ is the first and only commercially available graph-native ML functionality for enterprises, and we’re immensely happy to help democratize these innovations.
This release enables anyone to employ cutting edge ML techniques, which will change the way enterprises use their connected data.
Graph Embeddings
The graph embedding algorithms are the star of the show in this release.
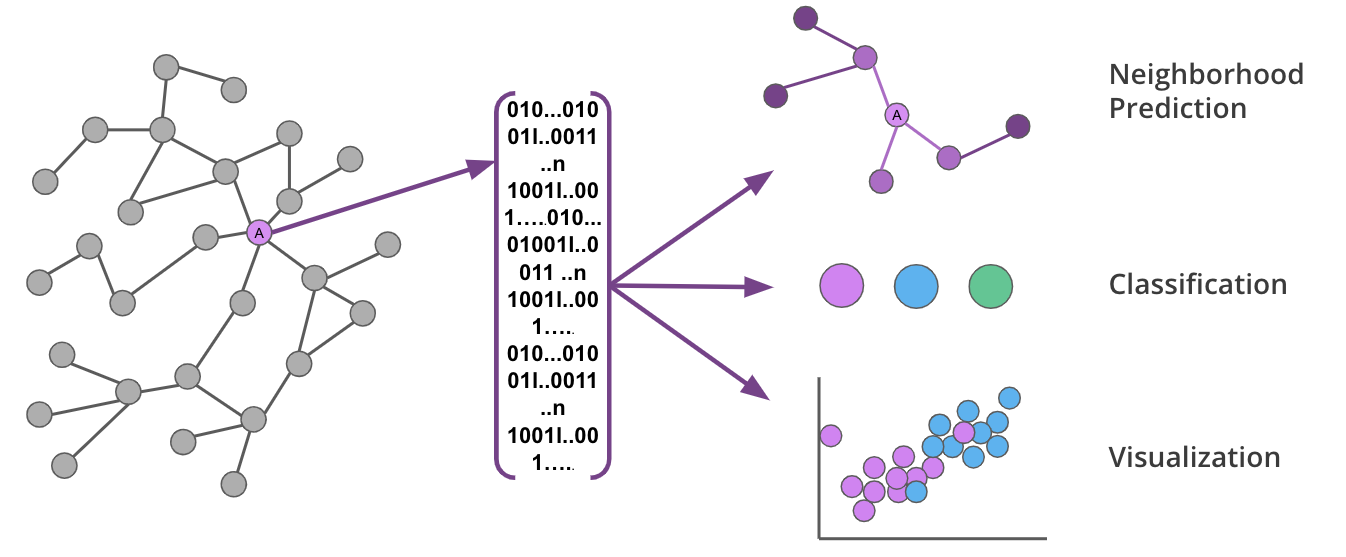
These algorithms are used to transform the topology and features of your graph into fixed-length vectors (or embeddings) that uniquely represent each node.
Graph embeddings are powerful, because they preserve the key features of the graph while reducing dimensionality in a way that can be decoded. This means you can capture the complexity and structure of your graph and transform it for use in various ML predictions.
Graph embeddings capture the nuances of graphs in a way that can be used to make predictions or lower dimensional visualizations.
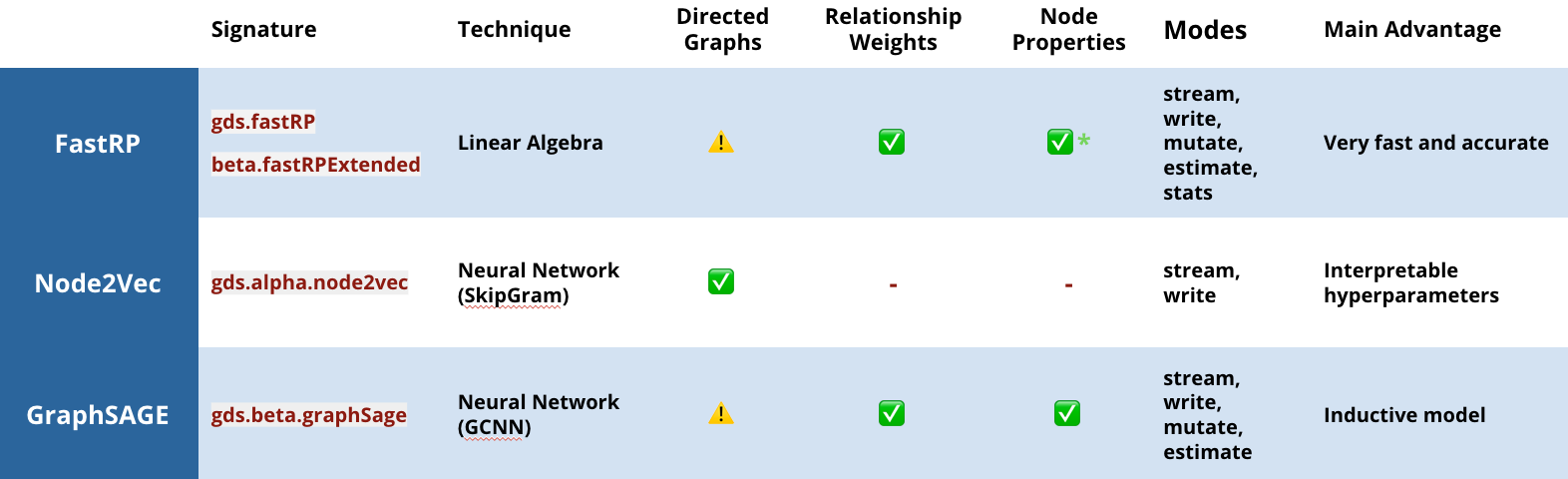
In this release, we are offering three embedding options that learn the graph topology and, in some cases, node properties to calculate more accurate representations:
- This is probably the most well-known graph embedding algorithm. It uses random walks to sample a graph, and a neural network to learn the best representation of each node.
- A more recent graph embedding algorithm that uses linear algebra to project a graph into lower dimensional space. In GDS 1.4, we’ve extended the original implementation to support node features and directionality as well.
- FastRP is up to 75,000 times faster than Node2Vec, while providing equivalent accuracy!
- This is an embedding technique using inductive representation learning on graphs, via graph convolutional neural networks, where the graph is sampled to learn a function that can predict embeddings (rather than learning embeddings directly). This means you can learn on a subset of your graph and use that representative function for new data and make continuous predictions as your graph updates. (Wow!)
- If you’d like a deeper dive into how it works, check out the GraphSAGE session from the NODES event.
Graph embeddings available in the Neo4j Graph Data Science Library v1.4 . The caution marks indicate that, while directions are supported, our internal benchmarks don’t show performance improvements.
Graph ML Model Catalog
GraphSAGE trains a model to predict node embeddings for unseen parts of the graph, or new data as mentioned above.
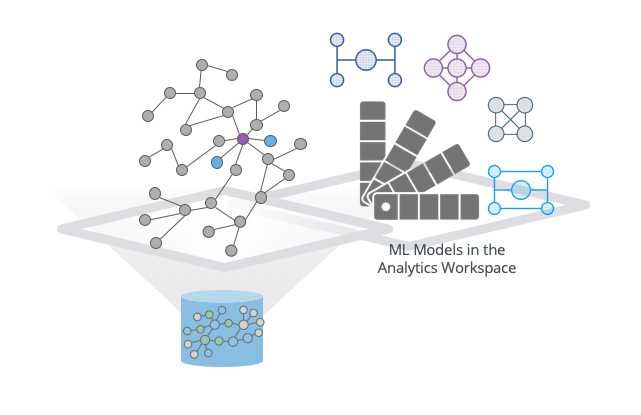
To really capitalize on what GraphSAGE can do, we needed to add a catalog to be able to store and reference these predictive models. This model catalog lives in the Neo4j analytics workspace and contains versioning information (what data was this trained on?), time stamps and, of course, the model names.
When you want to use a model, you can provide the name of the model to GraphSAGE, along with the named graph you want to apply it to.
GraphSAGE ML Models are stored in the Neo4j analytics workspace.
Yes, we now have our first ML models inside Neo4j! But so what? Let’s take a look at a real-world example.
Example: Knowledge Graph Completion for Drug Discovery
Knowledge graph completion is important to a range of industries because we rarely, if ever, have complete information about a complex system.
In life sciences, knowledge graph completion is essential for identifying new associations between genes and diseases, drug discovery and to repurposing existing drugs.
Let’s look at how Neo4j can be used for knowledge graph completion in the area of drug discovery:
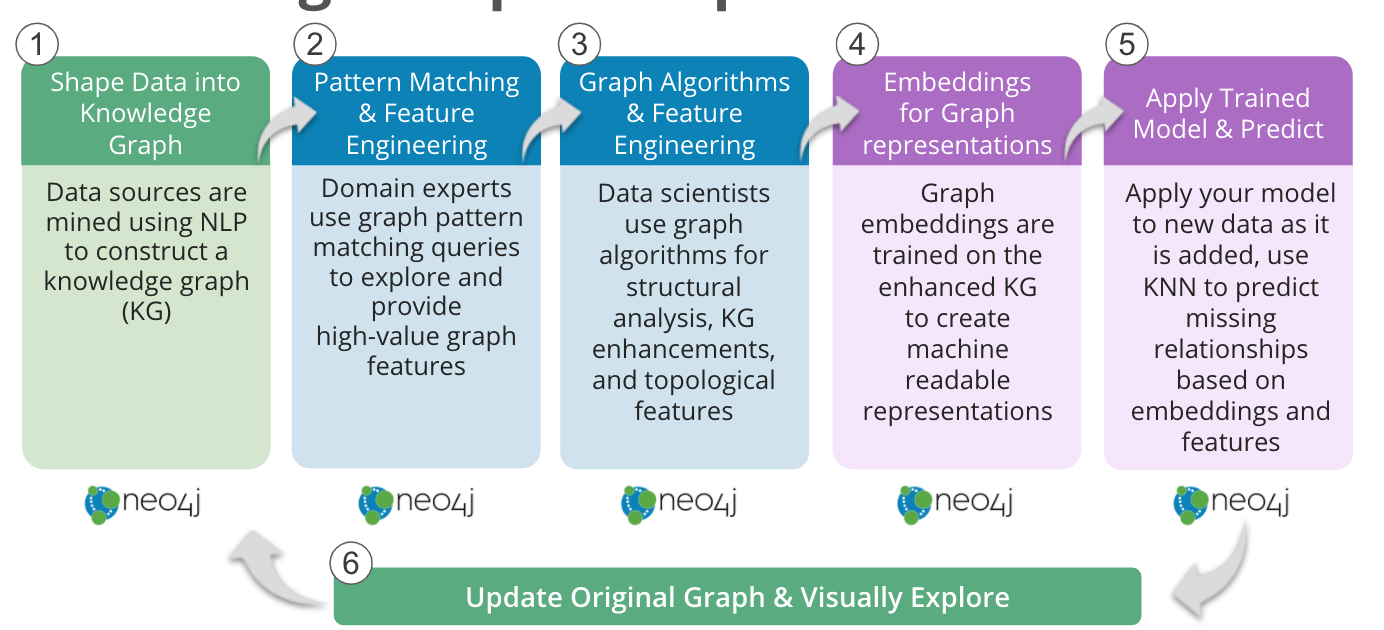
Knowledge Graph Completion Workflow in Neo4j
- Share Data into a Knowledge Graph
Academic publications, patents, medical records and laboratory data can be mined using natural language processing (NLP) techniques to construct a knowledge graph, such as COVIDgraph.org or Hetionet. - Pattern Matching and Feature Engineering
Domain experts can perform graph pattern matching using Cypher queries to find drugs reportedly effective for the genes related to a disease of interest, but not yet in use to treat a particular disease. Similarly, expert-driven queries can provide high-value graph features: For example, calculating probability scores for potential drugs based on graph pattern matching. - Graph Algorithms and Feature Engineering
Neo4j graph algorithms can be used to enrich the information available about the graph. For example, Centrality algorithms can reveal the importance of individual genes, Community Detection algorithms can identify regulatory networks, or PathFinding algorithms can reveal how closely associated a drug may be with a disease of interest. - Embeddings for Graph Representations
The graph embeddings in Neo4j can be trained on this more complete knowledge graph to learn the extended network topology, as well as incorporating the query and algorithm derived features, to create machine-readable representations of the knowledge graph. - Apply Trained Model and Predict
Machine learning graph database models can then be trained to predict, based on the embeddings and other features, where edges should be in the graph – either facts that were missing from the original data or associations that have not yet been made. In Neo4j, the k-NN algorithm can be used to create edges between nodes based on similar embeddings. Alternatively, Neo4j’s Python driver can also pull embeddings into an existing workflow. - Update Original Graph and Visually Explore
Finally, the newly predicted relationships can then be added back into the source graph and explored with Neo4j Bloom.
In a drug discovery scenario, this means not only identifying possible new associations between genes, diseases, drugs and proteins, but also providing the immediate context to assess the relevance or validity of discoveries.
From queries that support domain experts in uncovering what they know, to patterns that unearth and understand trends, to calculating high-value features that train ML models, knowledge graph completion isn’t possible without graph technology.
You can see why we’re so excited about this release that brings state-of-the-science ML techniques to graph-specific tasks, while also minimizing the number of tools you have to move in between. To get your hands on the latest from the GDS Library, visit our download center or go straight to our GitHub repo.
Finally, thank you to those who explored these algorithms in the alpha tier. We’re extremely grateful for your feedback. Please let us know what new features you’d like to see and how we can keep improving.
Cheers!
Alicia & Amy
Grab yourself a free copy of the Graph Data Science for Dummies book for an easy-to-understand guide to the basics of GDS – no previous background required.