How to Build a GenAI Chatbot From Technical Documents Using Neo4j and Unstructured.io
6 min read
A common challenge when building GenAI applications for technical audiences is providing accurate responses that also contain relevant unstructured information, such as charts, diagrams, and tables.
In this blog, we’ll build a simple GenAI chatbot using open-source technical documentation from the energy industry. We’ll use Unstructured.io to perform high-resolution chunking, extracting tables and images along the way, and build a Neo4j knowledge graph that preserves the chunk sequence in document context. We’ll deploy a chatbot built using Neo4j’s Needle Starter Kit, which will perform inspectable retrieval and generation operations with the Neo4j GraphRAG Package for Python (neo4j-graphrag). Later, we’ll step through how to:
- Build vector indexes on chunks and full-text indexes on entity terms
- Parse and ingest document text, images, and tables into the knowledge graph
- Perform vector embedding on document chunks
- Perform entity extraction on document chunks
- Add metadata to the images to filter out junk images
- Deploy the chatbot application, using the neo4j-graphrag HybridCypherRetriever to perform simultaneous semantic and full-text chunk discovery, augmented by nearest-neighbor traversals to related chunks, images, and tables
But first, let’s see what the finished result looks like and why we’re going down this path.
A Chatbot That Understands Your Documents
Let’s start with the knowledge graph schema. :Document nodes have :Chunk nodes, these are sequenced in document order with a :NEXT_CHUNK relationship. :Chunk nodes have an embedding property for the vector backed by a Neo4j vector index. Images and tables in the text chunk are saved as :Image and :Table nodes, and kept in context with a :RELATED_CONTENT relationship. Extracted entities are instantiated as :Entity nodes (backed by a Neo4j Lucene full-text index) and mapped to chunks (within and across documents) by the :HAS_ENTITY relationship. This is known as a lexical graph, since it has decomposed the document into its various elements.
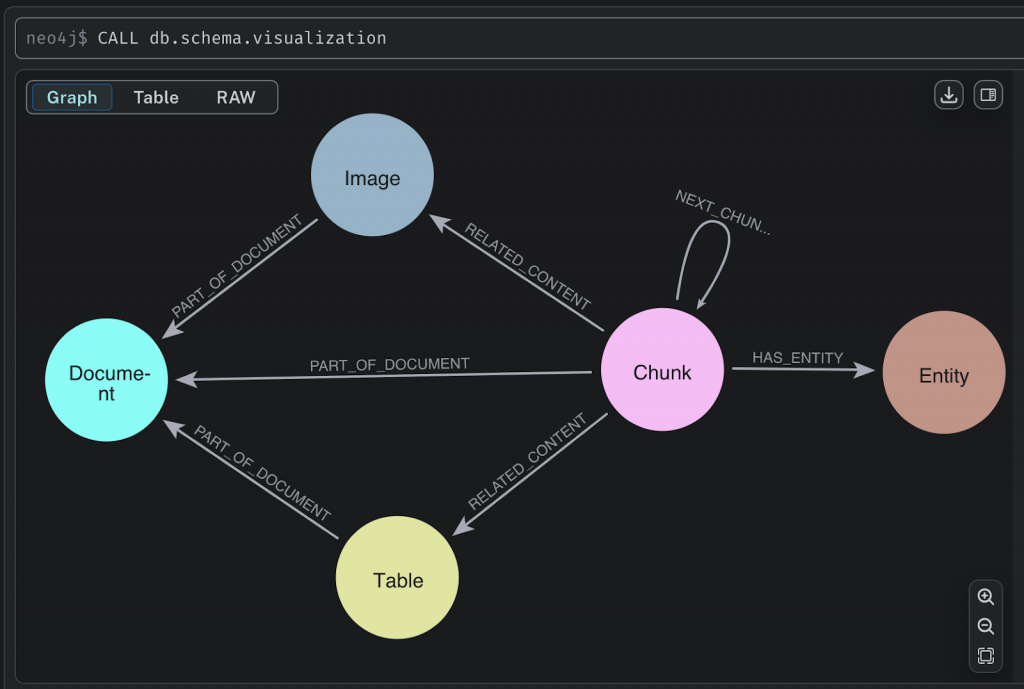
We can take a look at how the document is represented in Neo4j using this query:
MATCH path0 = (n:Document {name: "StatoilHydro_Volve_15_9_F_14_Report.pdf"})<-[:PART_OF_DOCUMENT]-(c0:Chunk)
WHERE NOT (c0)<-[:NEXT_CHUNK]-()
MATCH (n)<-[:PART_OF_DOCUMENT]-(c:Chunk)
WITH path0, COLLECT(c) AS chunks
UNWIND chunks AS chunk
CALL(chunk) {
MATCH path1 = (chunk)-[:NEXT_CHUNK|RELATED_CONTENT]-()
RETURN path1
}
RETURN path0, path1
The graph preserves the basic structure of the document – a sequence of :Chunk nodes chained in order of appearance, along with any associated :Image and :Table nodes. For this particular document, we have parsed 47 narrative text chunks, 32 images, and 11 tables.
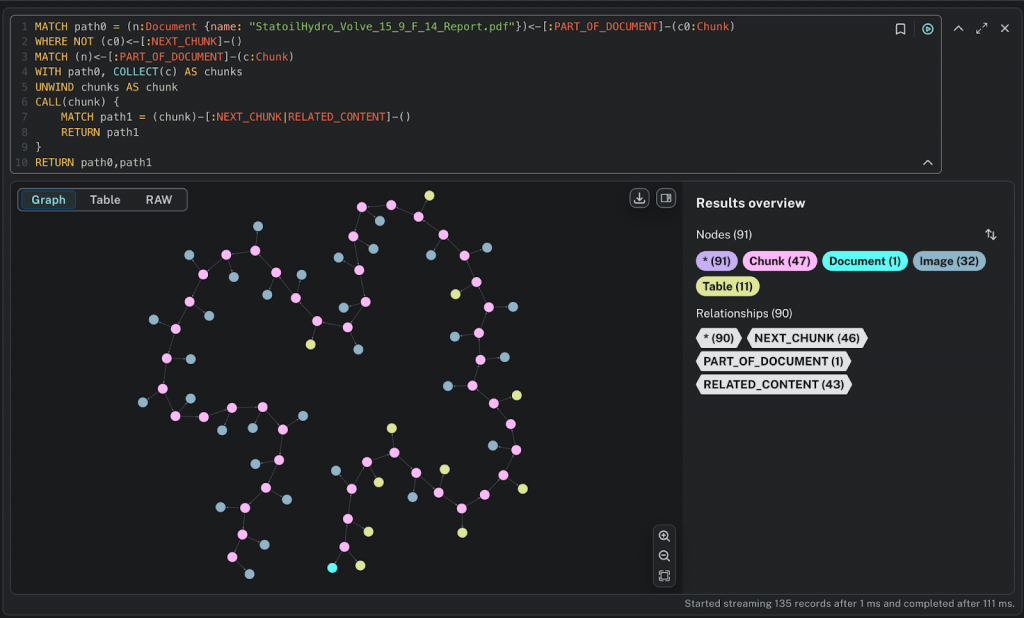
We can zoom in and see how this looks. The :Document node is in the lower left corner, and you can see the sequenced :Chunk nodes with related :Image and :Table nodes.
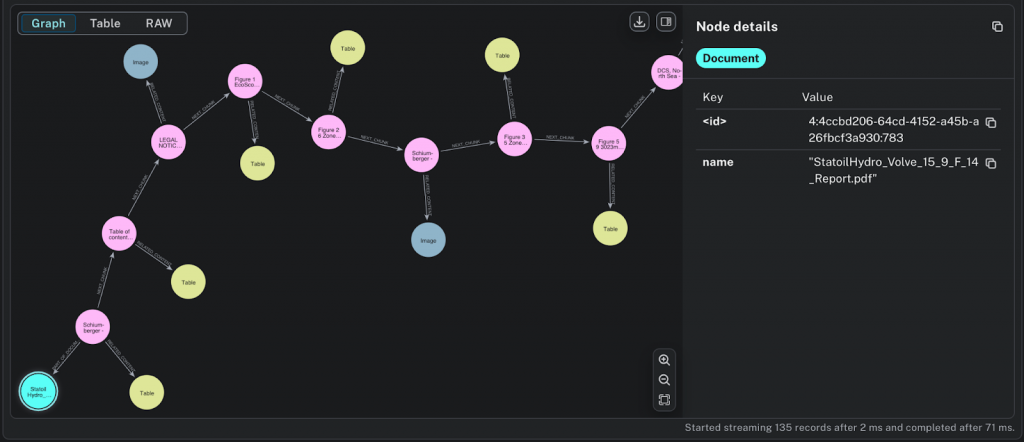
:Document, :Chunk, :Image, and :Table nodesAccuracy and Explainability With Neo4j GraphRAG
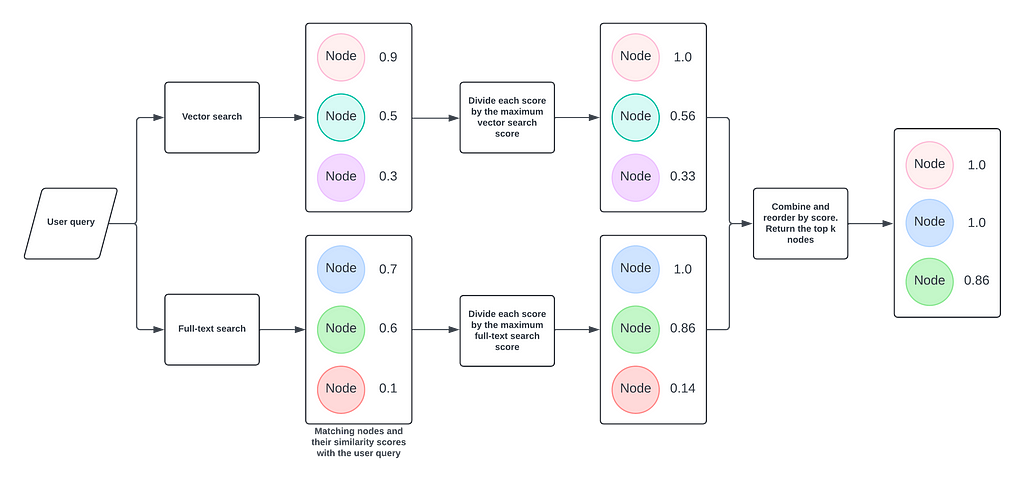
So how does this help us improve GenAI accuracy? With this lexical graph structure, we can discover texts (and related content) using four retrieval methods:
- Semantic search of
:Chunktext embeddings using vector indexing - Full-text index search of extracted entities and traversal to their source texts
- Traversal to nearest-neighbor texts for
:Chunknodes discovered by methods 1 and 2 - Traversal to images and tables related to
:Chunknodes discovered by methods 1 and 2
Results for methods 1-3 are packaged to provide rich, accurate, and structured context for summarization by the LLM. Results for method 4 (images and tables) are displayed post-generation in the chatbot (see below).
This all sounds complicated, but fortunately, the heavy lifting is done by the HybridCypherRetriever method in the neo4j-graphrag package:
#this query pulls the adjacent Chunks, Entities
RETRIEVAL_QUERY = (
"""
WITH node, score
OPTIONAL MATCH (node)-[:NEXT_CHUNK]-(c) // get chunk neighbors
OPTIONAL MATCH (node)<-[HAS_ENTITY]-(e) // get entity context chunks
ORDER BY score DESC LIMIT 100
RETURN apoc.convert.toSet(COLLECT(elementId(node))+COLLECT(elementId(e))+COLLECT(elementId(c))) AS listIds,
COLLECT (e.id) as contextNodes, node.text as nodeText, score ORDER BY score DESC
"""
)
def __init__(self, driver, embedder, vector_index_name, fulltext_index_name):
self._retriever = HybridCypherRetriever(
driver,
vector_index_name=vector_index_name,
fulltext_index_name=fulltext_index_name,
retrieval_query=self.RETRIEVAL_QUERY,
result_formatter=self.formatter,
embedder=embedder,
neo4j_database="neo4j",
)
We tell the retriever the names of the vector and full-text indexes to use, pass in the embedder for the user’s prompt, and specify the augmenting Cypher query we’re using to traverse to nearest-neighbor :Chunk nodes from the discovered nodes, along with the database to use.
Example Prompt
Let’s see it in action. There are some example questions in the repo, so let’s try one:
What is meant by a sinusoid in the EcoScope analyses?
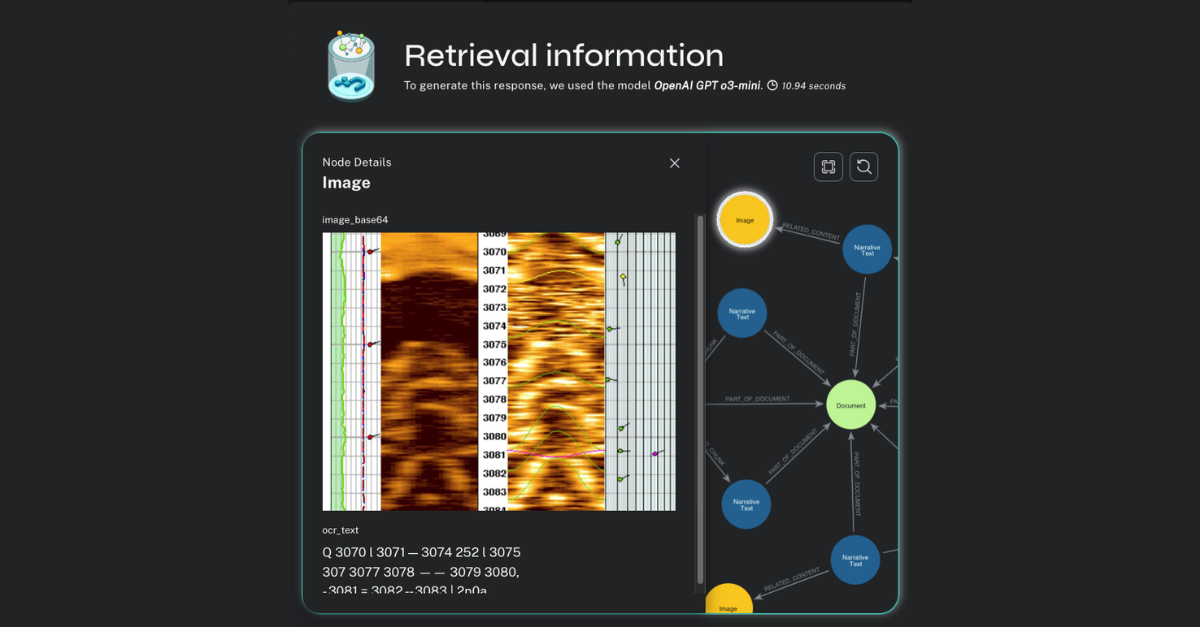
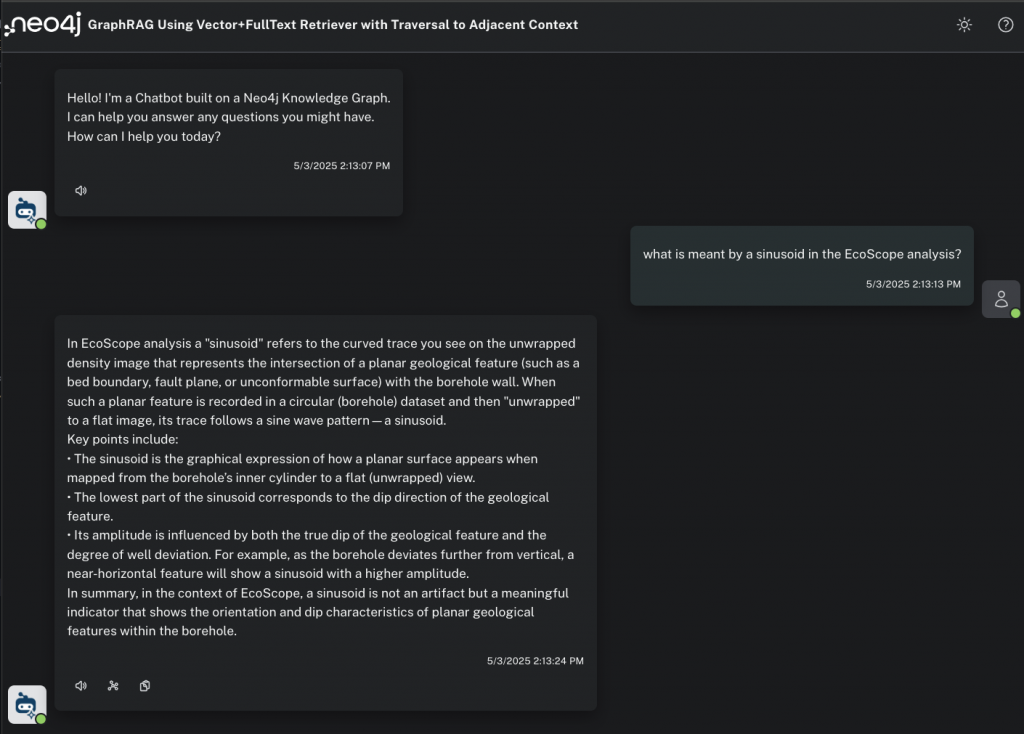
We get a robust, specific answer back (and as a former earth scientist, having read these documents closely, I can say this is a good and correct result), but let’s see if we’re really convinced. Pretending to be a petroleum engineer, I can click on the graph icon to inspect the content used to prepare the response.
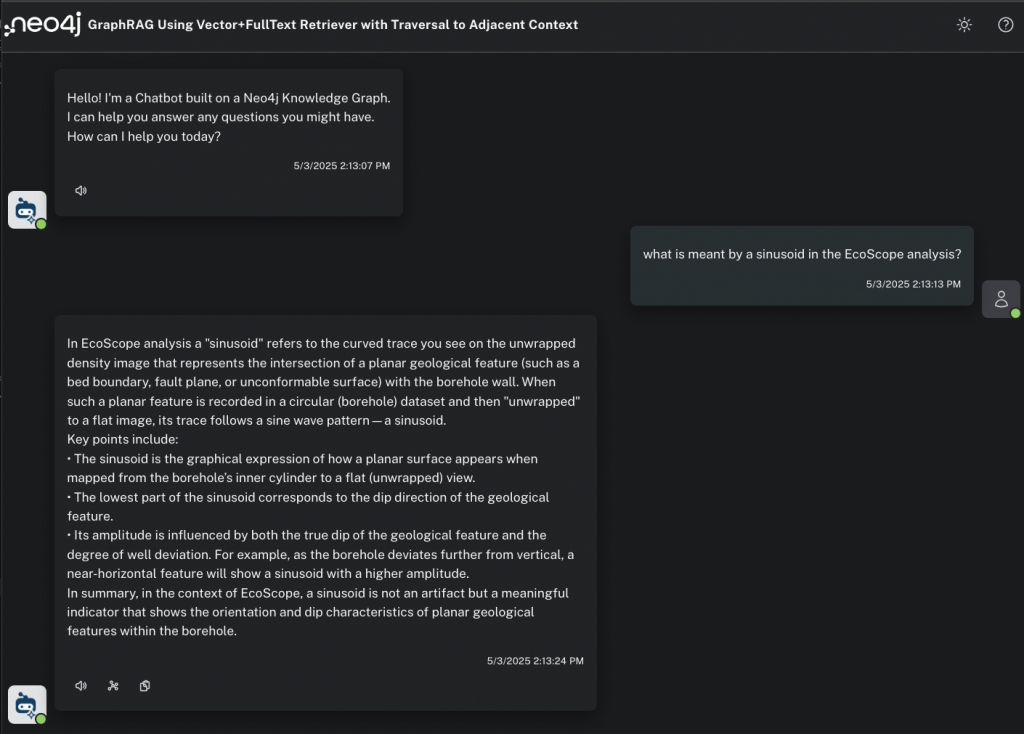
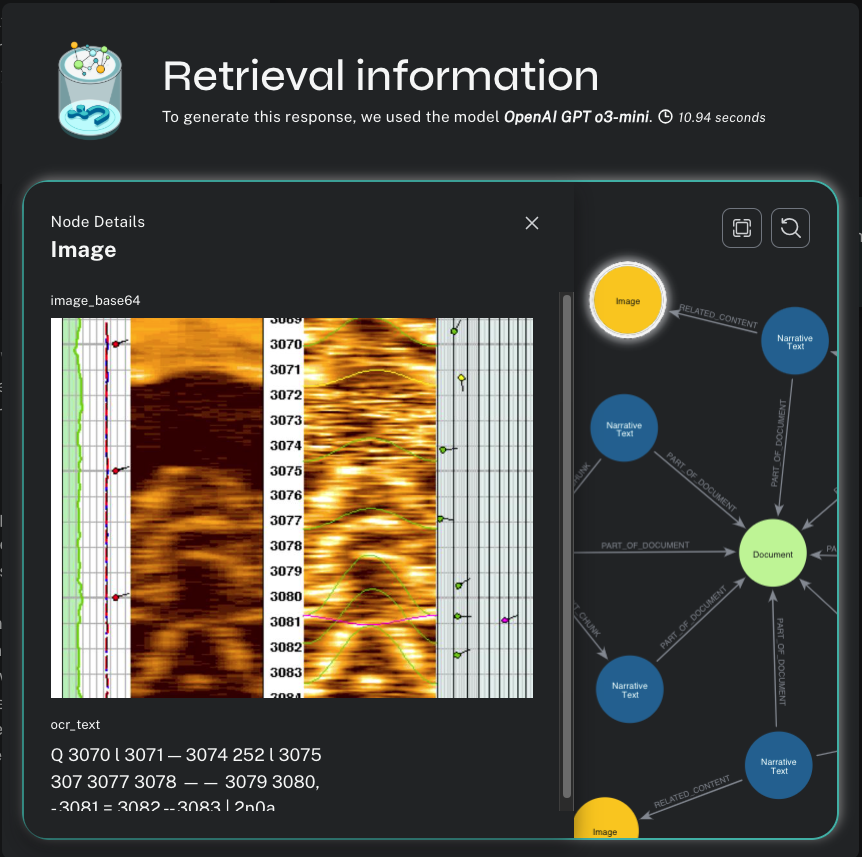
Now we can see the inner workings of the HybridCypherRetriever. We have a lexical subgraph to explore that shows the :Chunk and :Entity nodes discovered by semantic and full-text search, respectively; the nearest-neighbor chunks discovered by local traversal inside the document sequence; and all the related :Table and :Image nodes.
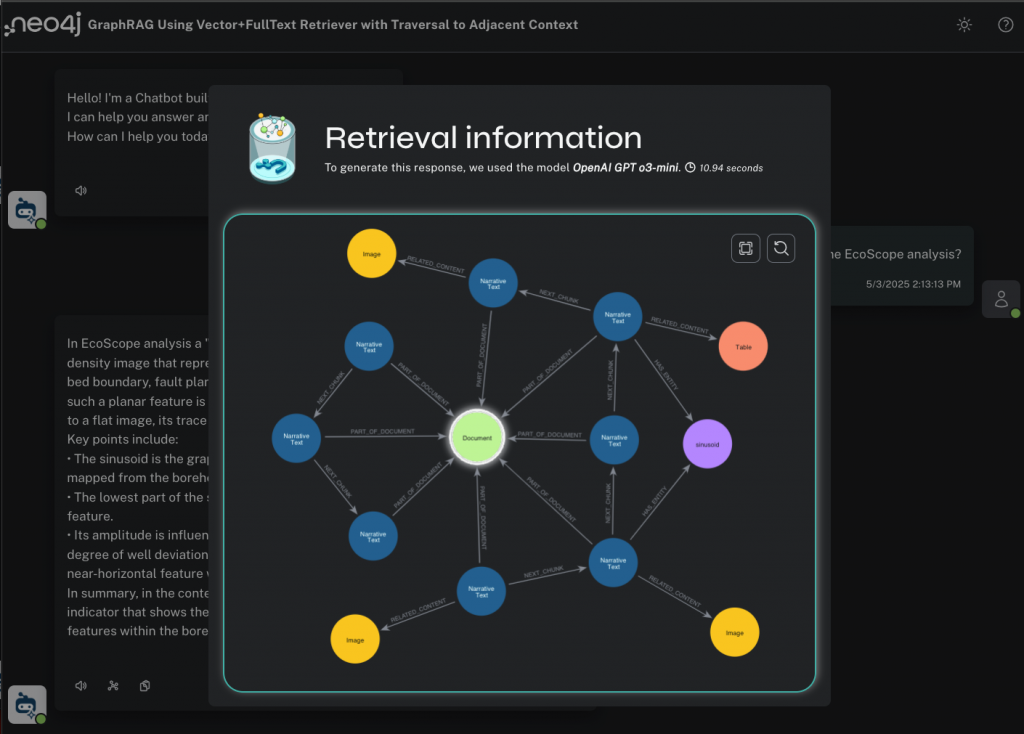
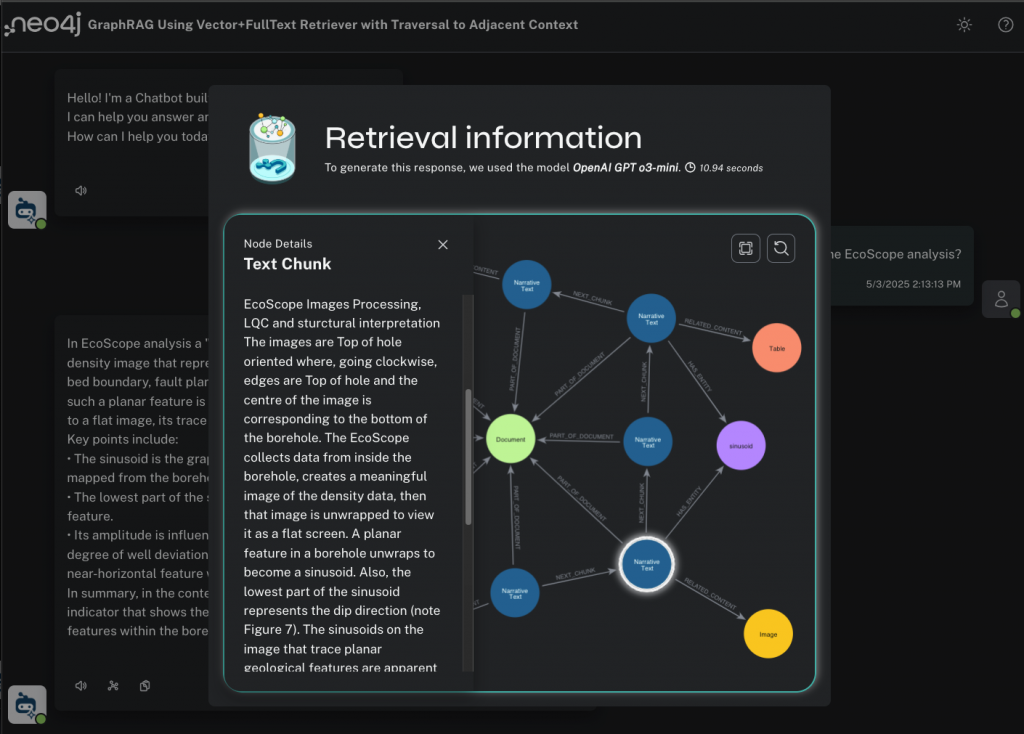
I can click on any of these nodes in the visualization and see their content.
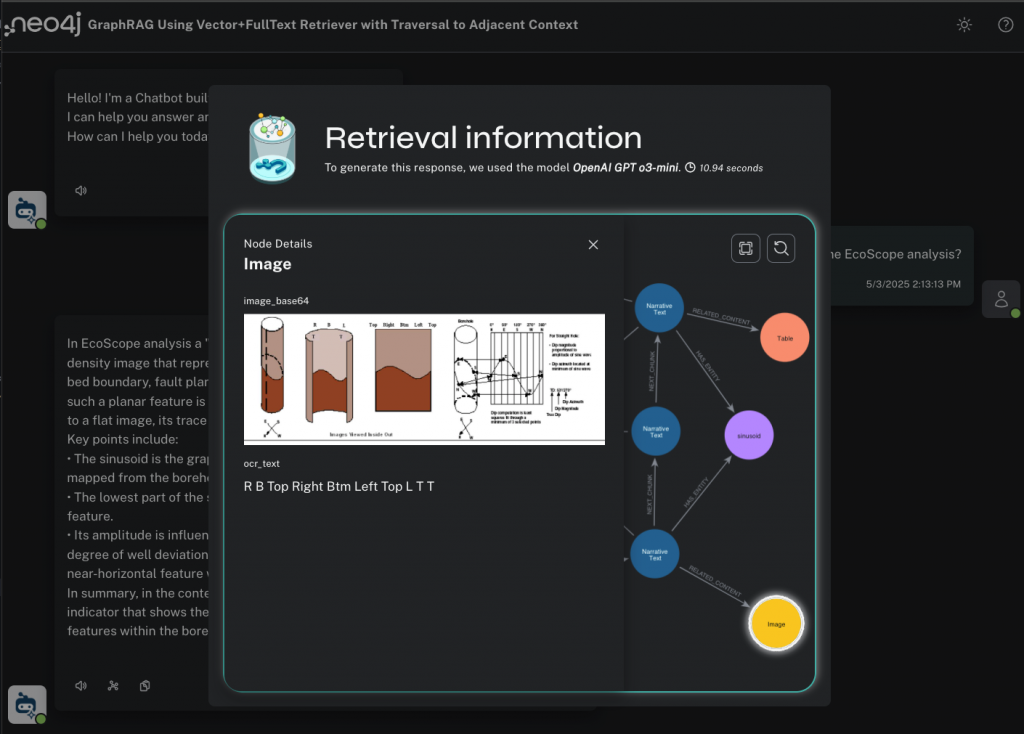
And if I click on an :Image node, I can see a diagram that shows how the EcoScope analysis works, which might be the best and most intuitive answer to my original question.
So What? The GenAI Quick-Take
This blog shows how Neo4j knowledge graphs can provide contextualized, accurate, and inspectable GenAI responses (complete with images and tables) for technical audiences. Achieving this level of specificity and trust is challenging using only LLMs or vector databases.
Unstructured.io enables intelligent extraction of narrative text in document context, along with associated images and tables for additional explanatory context. Neo4j enables you to easily build a knowledge graph that combines both descriptive information from unstructured data and business facts & hierarchy from structured data.
By leveraging a Neo4j knowledge graph as the persistence layer for your GenAI application, you can provide a performant and trustworthy experience for your end users, layering on agentic frameworks and MCP integrations as needed for even more sophisticated knowledge discovery methods.
The concepts presented here can be extended to any corpus of technical documents with only slight modifications to the repo code. The next sections step through how you can leverage these approaches to build a Neo4j knowledge graph and chatbot from your own documents.
Document Parsing With Unstructured.io
First, we need to parse our technical documents. To do this, we’ll use some great tools from Unstructured.io, a new Neo4j partner.
Unstructured.io provides sophisticated tooling for extracting a wide range of content elements from documents and is able to examine the document layout to infer additional context. Unstructured.io recently released a Neo4j connector that allows you to build a lexical graph of text chunks as part of a parallelized Unstructured.io document processing pipeline. You can use Unstructured.io services through their web console or API.
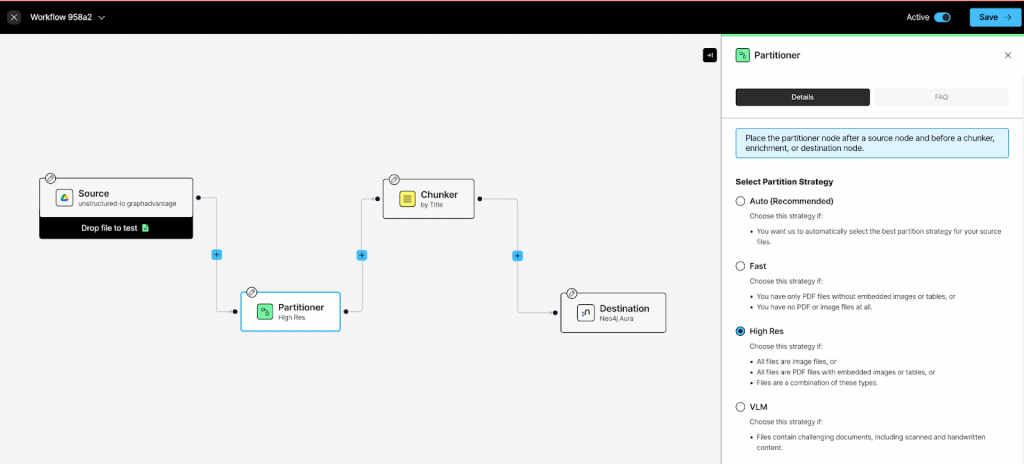
Unstructured.io pipelines use a few basic components. We’ll focus on the partitioner, chunker, embedder, entity extractor, and Neo4j destination.
The partitioner is the workhorse of the Unstructured.io platform. Able to operate in parallel, it can quickly shred a document, then identify and tag elements like titles, headers, footers, page numbers, narrative text, lists, formulas, etc. – and, as we’ll see later, even extract images and tables.
partitioner_config=PartitionerConfig(
partition_by_api=True,
api_key=(UNSTRUCTURED_API_KEY),
partition_endpoint=UNSTRUCTURED_API_URL,
additional_partition_args={
"split_pdf_page": True,
"split_pdf_allow_failed": True,
"split_pdf_concurrency_level": 15
}
The chunker is a configurable layer of contextual intelligence that gathers elements produced by the partitioner into larger text blocks for presentation to an LLM. If you use the by_title option, it’ll look at document indenting and whitespace to determine natural boundaries in the narrative.
chunker_config=ChunkerConfig(
chunking_strategy="by_title"
max_characters=1500
),
The embedder generates vector embeddings for chunks to support semantic search, and the entity extractor parses entities from the chunks and can pass a prompt to the LLM.
embedder_config=EmbedderConfig(
embedding_provider="openai",
embedding_api_key=OPENAI_API_KEY
),
The Neo4j destination connector builds some constraints and native vector indexes, then loads the pipeline’s output into Neo4j, building a lexical graph that connects sequenced chunk nodes to a node representing the processed document. Entity nodes connect chunk nodes for additional traversable context.
destination_connection_config=Neo4jConnectionConfig(
access_config=Neo4jAccessConfig(password=NEO4J_PASSWORD),
username=NEO4J_USERNAME,
uri=NEO4J_URI,
database=NEO4J_DATABASE,
),
stager_config=Neo4jUploadStagerConfig(),
uploader_config=Neo4jUploaderConfig(batch_size=100)
For our application, we’ll use the Unstructured.io parsing API, performing all the parsing and chunking operations. The GitHub repo contains all the notebooks, application code, and links to example files. Follow the repo instructions on how to provision a free Neo4j Aura graph database and get your Unstructured.io API key.
I’ll go through building the graph step by step in the next section.
Lexical Graph Pipeline
Here’s the full pipeline used to build the lexical knowledge graph.
Step 1. Build Vector Indexes on Document Chunks and Full-Text Indexes on Entity Terms
First, we’ll do some configurations and set up the necessary indexes for native semantic and full-text searching in Neo4j using the neo4j-graphrag package.
Install Dependencies
!pip install unstructured-client !pip install neo4j !pip install neo4j-graphrag
Configure Variables
unstructured_api_key="UNSTRUCTURED_API_KEY" neo4j_uri = "NEO4J_URI" neo4j_database = "neo4j" neo4j_user = "neo4j" neo4j_password = "NEO4J_PASSWORD" openai_api_key ="OPENAI_API_KEY"
Set Vector and Full-Text Indexes in Neo4j
from neo4j import GraphDatabase
from neo4j_graphrag.indexes import create_vector_index
from neo4j_graphrag.indexes import create_fulltext_index
# Neo4j driver setup
driver = GraphDatabase.driver(neo4j_uri, auth=(neo4j_user, neo4j_password))
VECTOR_INDEX_NAME = "chunk_embedding"
# Creating the vector index
create_vector_index(
driver,
VECTOR_INDEX_NAME,
label="Chunk",
embedding_property="embedding",
dimensions=1536,
similarity_fn="cosine",
fail_if_exists=False,
)
FULLTEXT_INDEX_NAME = "entity_text"
# Creating the full text index
create_fulltext_index(
driver,
FULLTEXT_INDEX_NAME,
label="Entity",
node_properties= ["text", "variants"],
fail_if_exists=False,
)
query = '''
SHOW INDEXES
'''
with driver.session() as session:
result = session.run(query)
for record in result:
print(record)
Step 2. Parse and Ingest Document Text, Images, and Tables Into the Knowledge Graph
Next, we’ll parse our document set and build the knowledge graph. We’ll create nodes for :Document, :Chunk, :Image, and :Table. You’ll see that we use the apoc.nodes.link() procedure to link all of the :Chunk nodes in order of appearance in the :Document, and also that we’re creating additional :Image and :Table nodes if they were part of the :Chunk content. We’re storing the images as binaries and OCR text, and tables as images, HTML, and text.
Extract and Chunk PDFs With Unstructured.io
import os
import base64
import zlib
import json
import logging
import nltk
from neo4j import GraphDatabase
from unstructured_client import UnstructuredClient
from unstructured_client.models import operations, shared
from unstructured.staging.base import elements_from_dicts, elements_to_json
# Disable logging
logging.disable(logging.CRITICAL)
# Configuration
directory_path = "PATH_TO_DOCUMENTS/"
client = UnstructuredClient(
api_key_auth=unstructured_api_key,
server_url="https://api.unstructuredapp.io"
)
driver = GraphDatabase.driver(neo4j_uri, auth=(neo4j_user, neo4j_password))
CHUNK_QUERY = '''
WITH apoc.convert.fromJsonList($json) AS maps
UNWIND maps AS map
WITH apoc.map.clean(map,[],[" ",""]) AS m
MERGE (d:Document {name: m.metadata.filename})
WITH m, d
CALL(m, d) {
CREATE (n:Chunk {id: m.element_id})
SET
n.type = "NarrativeText",
n.text = m.text,
n.filename = m.metadata.filename,
n.filetype = m.metadata.filetype,
n.languages = m.metadata.languages,
n.page_number = m.metadata.page_number,
n.tokens = m.tokens
CREATE (n)-[:PART_OF_DOCUMENT]->(d)
RETURN n
}
WITH m, d, n
CALL(m, d, n) {
WITH m, d, n
WHERE m.metadata.type IN ["Image", "Table"]
CREATE (i:$(m.metadata.type) {id: m.element_id})
SET i.type = m.metadata.type,
i.figure_caption = m.metadata.figure_caption,
i.text = m.metadata.text,
i.filename = m.metadata.filename,
i.filetype = m.metadata.filetype,
i.languages = m.metadata.languages,
i.page_number = m.metadata.page_number,
i.image_base64 = m.metadata.image_base64,
i.image_mime_type = m.metadata.image_mime_type,
i.text_as_html = m.metadata.text_as_html
MERGE (n)-[:RELATED_CONTENT]->(i)
MERGE (i)-[:PART_OF_DOCUMENT]->(d)
}
WITH DISTINCT d, n
WITH d, COLLECT(n) AS nodes
CALL apoc.nodes.link(nodes, "NEXT_CHUNK")
'''
def run_query(tx, query, json_data):
return tx.run(query, {"json": json_data}).consume()
def extract_orig_elements(encoded):
decoded = base64.b64decode(encoded)
decompressed = zlib.decompress(decoded)
return json.loads(decompressed.decode("utf-8"))
def process_file(filepath, filename):
print(f"\nProcessing file: {filename}")
with open(filepath, "rb") as f:
files = shared.Files(
content=f.read(),
file_name=filename
)
request = operations.PartitionRequest(
partition_parameters=shared.PartitionParameters(
files=files,
strategy="hi_res",
hi_res_model_name="yolox",
element_exclude=["Header", "Footer", "ListItem", "Formula", "UncategorizedText"],
extract_image_block_types=["Image", "Table"],
chunking_strategy="by_title",
max_characters=1500,
split_pdf_page=True,
split_pdf_allow_failed=True,
split_pdf_concurrency_level=15
)
)
response = client.general.partition(request=request)
element_dicts = [e for e in response.elements]
for i, element in enumerate(element_dicts):
if element.get("text"):
element["tokens"] = len(nltk.word_tokenize(element["text"]))
metadata = element.get("metadata", {})
if metadata.get("orig_elements"):
orig_elements = extract_orig_elements(metadata["orig_elements"])
for obj in orig_elements:
if obj.get("type") == "FigureCaption" and obj.get("text", "").lower().startswith("figure"):
metadata["figure_caption"] = obj["text"]
if obj.get("type") == "Image":
metadata.update({
"element_id": obj["element_id"],
"type": obj["type"],
"image_base64": obj["metadata"]["image_base64"],
"image_mime_type": obj["metadata"]["image_mime_type"],
"text": obj["text"]
})
if obj.get("type") == "Table":
metadata.update({
"element_id": obj["element_id"],
"type": obj["type"],
"text_as_html": obj["metadata"]["text_as_html"],
"image_base64": obj["metadata"]["image_base64"],
"image_mime_type": obj["metadata"]["image_mime_type"],
"text": obj["text"]
})
element_dicts[i]["metadata"].pop("orig_elements", None)
json_data = json.dumps(element_dicts, indent=4)
with driver.session() as session:
summary = session.execute_write(run_query, CHUNK_QUERY, json_data)
print(f"nodes created => {summary.counters.nodes_created}, rels created => {summary.counters.relationships_created}")
session.close()
print(f"Finished processing: {filename}")
def main():
for filename in os.listdir(directory_path):
if filename.startswith(".") or not os.path.isfile(os.path.join(directory_path, filename)):
continue
try:
process_file(os.path.join(directory_path, filename), filename)
except Exception as e:
print(f"Error processing {filename}: {e}")
driver.close()
print("Done!")
if __name__ == "__main__":
main()
Step 3. Perform Vector Embedding on Document Chunks
Next, we’ll use OpenAI to perform vector embedding on the chunk texts and write these as properties. Because we already declared a vector index on the embedding property, similarities will be automatically calculated behind the scenes.
Embed Vectors With Neo4j GraphRAG
# vector embedding on Chunk text for semantic search
from neo4j import GraphDatabase
from neo4j_graphrag.embeddings import OpenAIEmbeddings
import logging
import sys
# Disable logging output
logging.disable(sys.maxsize)
# --- Configuration ---
EMBEDDING_MODEL = "text-embedding-ada-002"
MAX_CHUNK_LENGTH = 12000
# Initialize OpenAI embedder
embedder = OpenAIEmbeddings(model=EMBEDDING_MODEL, api_key=openai_api_key)
# Initialize Neo4j driver
driver = GraphDatabase.driver(neo4j_uri, auth=(neo4j_user, neo4j_password))
def embed_chunks():
with driver.session() as session:
result = session.run("""
MATCH (n:Chunk)
WHERE n.text IS NOT NULL AND n.embedding IS NULL
RETURN n.id AS id, n.text AS data
""")
for record in result:
chunk_id = record["id"]
data = record["data"]
if len(data) > MAX_CHUNK_LENGTH:
print(f"Skipping chunk {chunk_id} (length: {len(data)})")
continue
print(f"\rEmbedding chunk: {chunk_id}", end="", flush=True)
try:
embedding = embedder.embed_query(data)
session.run("""
MATCH (n:Chunk {id: $chunk_id})
SET n.embedding = $embedding
""", chunk_id=chunk_id, embedding=embedding)
except Exception as e:
print(f"\nFailed to embed chunk {chunk_id}: {e}")
def main():
embed_chunks()
driver.close()
print("\nDone embedding chunks!")
if __name__ == "__main__":
main()
Step 4. Perform Entity Extraction on Document Chunks
We’ll use OpenAI to perform entity extraction on the chunk texts and link the entities together. Because this is a compute-intensive operation, I like to use a :ProcessMe label to speed up the query.
You’ll notice that we’re using a data domain-specific prompt to guide the entity extraction and result format:
Extract all the entities from the following text. Identify only entities, abbreviations and technical terms commonly used in the petroleum exploration, petroleum geology, reservoir analysis, and oil & gas production. Return entities in this format: ["entity1", "entity2"]
Be sure to customize this for your own data domain to properly extract entities.
Label Chunk Nodes for Processing
# entity extraction on Chunk text for full text search
from neo4j import GraphDatabase
# Neo4j driver setup
driver = GraphDatabase.driver(neo4j_uri, auth=(neo4j_user, neo4j_password))
# to perform selective entity extraction using "ProcessMe" label execute this query
PROCESS_ME ='''
MATCH (n:Chunk {type:"NarrativeText"})
WHERE NOT (n)-[:HAS_ENTITY]->() AND n.entities IS NULL
SET n:ProcessMe
'''
with driver.session() as session:
res = session.run(PROCESS_ME)
session.close()
print("done!")
Entity Extraction on Labeled Chunk Nodes
# prompted entity extraction with some very light entity resolution
import logging
import sys
from neo4j import GraphDatabase
from openai import OpenAI
# --- Config ---
MAX_CHUNKS = 1000
OPENAI_MODEL = "gpt-4o" # or "gpt-4"
# --- Logging ---
logging.basicConfig(level=logging.INFO, format="%(asctime)s - %(levelname)s - %(message)s")
logging.disable(sys.maxsize) # Disable logging if needed
# --- Clients ---
client = OpenAI(api_key=openai_api_key)
driver = GraphDatabase.driver(neo4j_uri, auth=(neo4j_user, neo4j_password))
# --- Cypher Queries ---
FETCH_CHUNKS_QUERY = """
MATCH (n:Chunk:ProcessMe)
RETURN n.id AS id, replace(n.text,"\n","") AS text
LIMIT $limit
"""
ENTITY_INSERT_QUERY = """
WITH $entities AS entities
MATCH (n:Chunk:ProcessMe {id: $id})
WITH n, entities
CALL apoc.do.when(
entities[0] = "[]" OR entities[0] STARTS WITH "The text provided does not contain",
"WITH n SET n.entities = 'failed' REMOVE n:ProcessMe RETURN 0 AS rels",
"WITH n, apoc.convert.fromJsonList(entities[0]) AS names
UNWIND names AS name
MERGE (e:Entity {text: toLower(name)})
ON CREATE SET e.variants = [name]
ON MATCH SET e.variants = apoc.convert.toSet(e.variants + [name])
MERGE (n)-[:HAS_ENTITY]->(e)
WITH DISTINCT n, COUNT(e) AS rels
REMOVE n:ProcessMe
RETURN rels",
{n: n, entities: entities}
) YIELD value
RETURN value
"""
FAIL_MARK_QUERY = """
MATCH (n:Chunk:ProcessMe {id: $id})
SET n.entities = "failed"
REMOVE n:ProcessMe
"""
# --- Entity Extraction Prompt ---
# modify as needed for your data domain
def extract_entities(text: str) -> list:
prompt = f"""
Extract all the entities from the following text.
Identify only entities, abbreviations and technical terms commonly used in the petroleum exploration, petroleum geology, reservoir analysis, and oil & gas production.
Return entities in this format: ["entity1", "entity2"]
Do not include any extra text or explanation.
Text: {text}
"""
messages = [
{"role": "system", "content": "You help extract entities from petroleum-related text."},
{"role": "user", "content": prompt}
]
response = client.chat.completions.create(
model=OPENAI_MODEL,
messages=messages,
max_tokens=500,
temperature=0
)
return [response.choices[0].message.content.strip()]
# --- Main Function ---
def main():
processed_count = 0
with driver.session() as session:
chunks = session.run(FETCH_CHUNKS_QUERY, limit=MAX_CHUNKS)
for chunk in chunks:
chunk_id = chunk["id"]
text = chunk["text"]
processed_count += 1
try:
entities = extract_entities(text)
result = session.run(ENTITY_INSERT_QUERY, id=chunk_id, entities=entities)
for record in result:
rels = record["value"]["rels"]
print(f"\rRelationships created: {rels} | Processed: {processed_count} ", end="", flush=True)
result.consume()
except Exception as e:
logging.warning(f"\nFailed to process chunk {chunk_id}: {e}")
session.run(FAIL_MARK_QUERY, id=chunk_id)
driver.close()
print("\nDone extracting entities!")
if __name__ == "__main__":
main()
Step 5. Add metadata to the images to filter junk images
We’re almost done! If you were to look at some of the images captured, you’d see a lot of non-interesting graphics like logos and other extraction artifacts. As a final step, we’ll capture the image metadata so we can filter out non-interesting images when we do retrieval.
Calculate Image Sizes and Dimensions
#filtering properties for the frontend to hide logos and other uninteresting images
import base64
from io import BytesIO
from PIL import Image
from neo4j import GraphDatabase
import json
driver = GraphDatabase.driver(neo4j_uri, auth=(neo4j_user, neo4j_password))
# Cypher query to fetch nodes with base64 images
QUERY_IMAGES= """
MATCH (n:Image|Table)
WHERE n.image_base64 IS NOT NULL AND n.bytes IS NULL
RETURN n.id AS id, n.image_base64 AS image_base64
"""
def get_image_properties(image_base64: str):
try:
image_data = base64.b64decode(image_base64)
with Image.open(BytesIO(image_data)) as image:
width, height = image.size
aspect_ratio = max(width / height, height / width) if width and height else None
return {
"bytes": len(image_base64),
"width": width,
"height": height,
"aspect_ratio": aspect_ratio
}
except Exception as e:
print(f"Error processing image: {e}")
return None
def update_image_properties(driver):
with driver.session() as session:
result = session.run(QUERY_IMAGES)
for record in result:
node_id = record["id"]
image_base64 = record["image_base64"]
props = get_image_properties(image_base64)
if props:
session.run(
"""
WITH apoc.convert.fromJsonMap($json) AS map
MATCH (n:Image|Table {id: $id})
SET n += map
""",
{"id": node_id, "json": json.dumps(props)}
)
if __name__ == "__main__":
update_image_properties(driver)
driver.close()
print("Image metadata updated.")
GraphRAG Chatbot
Now that we have our graph built, let’s wire up the chatbot. Follow the instructions in the GitHub repo.
The application is built using the Neo4j Needle Starter Kit accelerator, and includes a Python backend and a React front end with some native graph visualizations using the Neo4j Visualization Library. The back-end component we’ll focus on is retriever.py.
You were probably wondering how the semantic search and full-text search indexes were going to work together. The quick answer is the hybrid retrievers that are part of neo4j-graphrag. The HybridCypherRetriever takes the user’s prompt and uses it as an input for a simultaneous vector search (of text embeddings) and full-text search (of entity terms), then returns the combined list of discovered nodes, ranked using a weighted average of the respective scores. Check out Hybrid Retrieval for GraphRAG Applications Using the GraphRAG Python Package for details.
In our application, we’re using the HybridCypherRetriever, which allows us to add a Cypher statement for additional traversals. This is where we can mine the knowledge graph for additional context – in this case, discovering chunks that both precede and follow (in document sequence) our search-discovered chunks. These neighbor chunks provide useful narrative context for a more complete and accurate GenAI result.
#this query pulls the adjacent Chunks, Entities
RETRIEVAL_QUERY = (
"""
WITH node, score
OPTIONAL MATCH (node)-[:NEXT_CHUNK]-(c) // get chunk neighbors
OPTIONAL MATCH (node)<-[HAS_ENTITY]-(e) // get entity context chunks
ORDER BY score DESC LIMIT 100
RETURN apoc.convert.toSet(COLLECT(elementId(node))+COLLECT(elementId(e))+COLLECT(elementId(c))) AS listIds,
COLLECT (e.id) as contextNodes, node.text as nodeText, score ORDER BY score DESC
"""
)
def __init__(self, driver, embedder, vector_index_name, fulltext_index_name):
self._retriever = HybridCypherRetriever(
driver,
vector_index_name=vector_index_name,
fulltext_index_name=fulltext_index_name,
retrieval_query=self.RETRIEVAL_QUERY,
result_formatter=self.formatter,
embedder=embedder,
neo4j_database="neo4j",
)
On the front end, we have a component RetrievalInformation.tsx (launched by the little graph icon in the result window), which displays all the :Chunk nodes sent to the LLM for summarization. It also displays the :Image and :Table nodes related to the summarized result. Each of these can be individually inspected by the user. This inspectability improves confidence in the system – the end user can see the discovered texts and related content that contributed to the final LLM summarized result.
RetrievalInformation.tsx
function run() {
const formattedSources = sources.map((source) => `"${source}"`).join(',');
const query1 = `
MATCH (a:Chunk)-[r:PART_OF_DOCUMENT]->(b:Document)
WHERE elementId(a) in [${formattedSources}]
RETURN DISTINCT a,r,b
UNION
MATCH (a:Chunk)-[r:HAS_ENTITY|NEXT_CHUNK]-(b:Chunk|Entity)
WHERE elementId(a) in [${formattedSources}] AND elementId(b) in [${formattedSources}]
RETURN DISTINCT a,r,b
UNION
MATCH (a:Chunk)-[r:RELATED_CONTENT]-(b:Image|Table)
WHERE elementId(a) in [${formattedSources}] AND b.aspect_ratio < 10 AND b.bytes > 1024 * 9
RETURN DISTINCT a,r,b
LIMIT 500
`;
I like clicking around in the result subgraph, but you could just as easily display these items inline with the text result. You may have noticed that Unstructured.io is also providing an OCR text result for the images and HTML for the tables. Another potential enhancement would be to make these texts searchable as well.
Summary
There are lots of potential enhancements and integrations you can add to this GenAI chatbot application, and as we’ve pointed out, you can apply the knowledge graph pipeline to any corpus of documents with only slight modifications to the code. If you have any questions about how to get started on your Neo4j GraphRAG project, please feel free to reach out to us.