Full-Text Search in 197M Chemical Names Graph Database

Back End Developer at CytoSMART
6 min read

PubChem is a database with millions of chemical compounds. All these can be downloaded and put into your graph database as a basis for your project.
I downloaded 197M synonyms related to 57M compounds for the Open Measurement project.
Open Measurement
This project is being built as part of “Open Measurement”, a platform to share measurements of biological experiments with others. The challenge comes from the many different formats and structures people use. This makes it hard to find out if other people did a similar experiment to yours. In this blog, I will look at building a compound searcher. The goal is that different synonyms of the same compounds can be linked to each other. Techniques: Neo4j graph database, full-text search, Lucene, data wrangling
Loading the Data
The data is given in turtle (.ttl) format. For the synonyms, there are 18 files with each over 10M rows. Each row connects a synonym value (name as a string) and an ID (MD5 encoding of the name). Next to those are 11 turtle files that link synonyms to compounds. The first 10 files also have 10M rows with the synonym ID and the compound ID (PubChem ID). The data has a few challenges:
- Empty strings for synonyms (957 synonyms)
- Duplicates of synonyms (1,591,056 synonyms)
- Synonyms with different md5 encoding as their ID (5,865 synonyms)
- There are more synonyms than compounds linked to synonyms
- Neosemantics (n10s) is a great tool to load in RDF files like turtles, but these files are too big (for my computer)
To solve these I made a python script that read all synonyms, then deleted the MD5 mismatches, duplicates, and empty strings. It saved them as smaller CSV files and load those into neo4j with “LOAD CSV”. Because I already dropped the duplicates I can use “CREATE” instead of “MERGE” to save time. This process only takes 2 to 3 hours. This results in 197M synonyms. For the compounds, I do a similar thing. I take all compounds from the “synonym2compound” files and remove the duplicates. Using the same CSV trick this process takes 1 to 2 hours and results in 57M compounds. Both synonyms and compounds are constrained to be unique and indexed on ID.
Next come the connections between synonyms and compounds, loading them all in smaller CSV, using “MERGE” to be sure no doubles are introduced. This process is taking days, maybe weeks (not finished when I started to write this). This takes so long because of 2 factors, for every connection the synonym and compound need to be found. Given they are both in a binary tree those are log(197M)=28 and log(57M)=26 searches. This already makes 50 times more operations than creating the nodes, merging can slow it down a bit more but the biggest one is the fact that I only had space on my HDD, not SSD meaning the I/O operations are killing me here. Neo4j setting:
dbms.memory.heap.initial_size=2G
dbms.memory.heap.max_size=6G
dbms.memory.pagecache.size=2G
Data exploration
Synonyms can have a relation to multiple compounds. An example is “destim” which is related to compounds 9306 and 66124. These two compounds are stereoisomerism but the name refers to both of them. PS, this does NOT apply to the compound 12213639, which is also a stereoisomerism of destim but destim does not revert to this compound.
Not all synonyms have a relation to a compound. This can already be seen by the mismatch in synonym files (18), and synonym-to-compound files (11).
Synonyms have, besides a name, also a type. For instance, oxydrene has the type “PubChem depositor-supplied molecular entity name”. This is not yet used in this version of the database.
Compounds can have 2d and 3d similarity metrics to each other. This holds great potential for finding a cluster of similar chemicals.
Full-Text Search Query
The first part is a standard tutorial we create a full-text index, this will use Lucene for us in the background.
CREATE FULLTEXT INDEX synonymsFullText FOR (n:Synonym) ON EACH [n.name]
Basic Fuzzy Full-Text Query
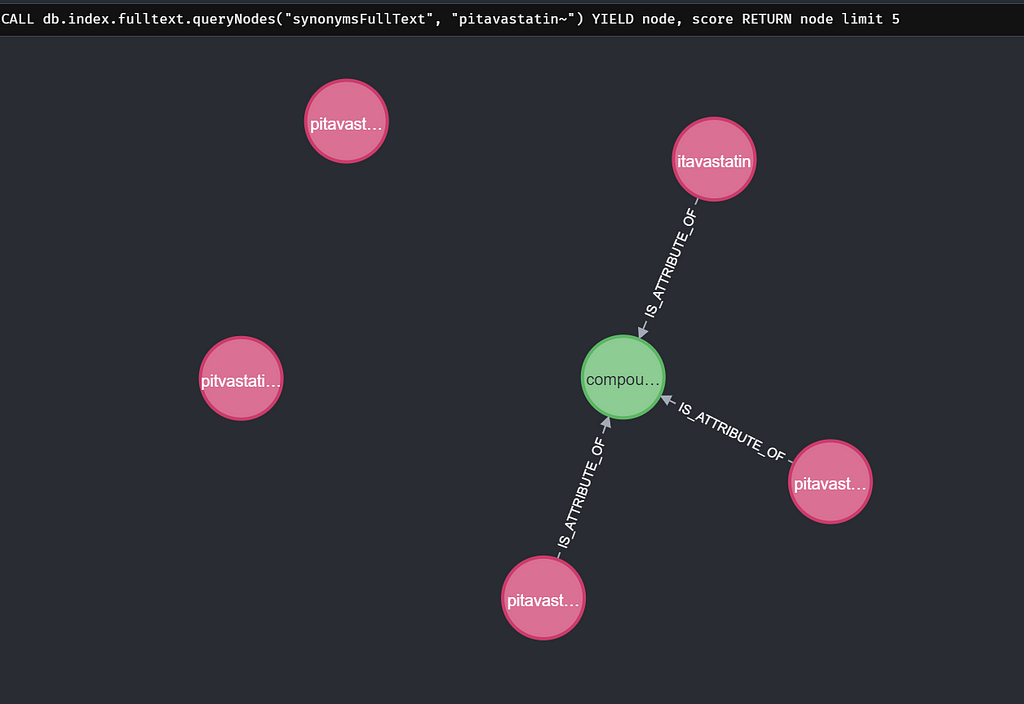
This means I can now use the Lucene query languages to find Synonym nodes by their name property. Lucene splits every index word into a given string. A normal search will return all nodes with an exact match of the word. When you add ~ after the word it will become a fuzzy match. Resulting in spelling mistakes being corrected. If I do this for “pitavastatan” while we mean pitavastatin it will work.
CALL db.index.fulltext.queryNodes("synonymsFullText", "pitavastatan~") YIELD node, score
RETURN node.name, score limit 5
One Synonym Per Compound Query
When I do this for “pitavastatin” we find 5 synonyms that match “pitavastatin” the best, but 3 of those are synonyms connected to the same compound. Meaning the autocomplete just gives you 3 unique compounds as options instead of 5. To solve this we are going to use the graph properties. We still are going to look for the synonyms but now take the compounds related to it. We only take one synonym per compound (the one with the highest full-text score). And then apply the limit of 5. This results in 5 unique compounds per search.
CALL db.index.fulltext.queryNodes("synonymsFullText", "pitavastatin~")
YIELD node, score
MATCH (node)-[:IS_ATTRIBUTE_OF]->(c:Compound)
WITH DISTINCT c as c, collect({score: score, node: node})[0] as s
RETURN s.node as name, s.score, c as compoundId limit 5
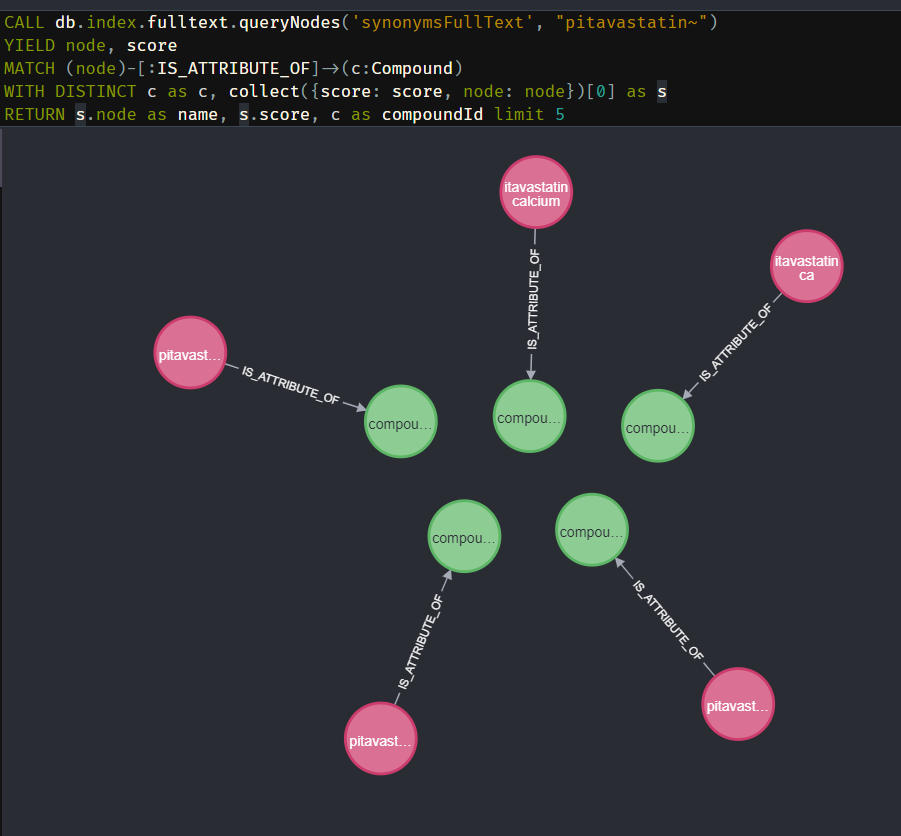
One Unique Synonym Per Compound Query
If a synonym has multiple compounds (like “destim”) it will return all compounds as a result. For example, destim will result in “destim cid9306” and “destim cid66124”. To combat this we will collect all the compounds related to the same synonym. Meaning the user will not have duplicates in their options. All related compounds are returned so users can be warned about it.
CALL db.index.fulltext.queryNodes("synonymsFullText", "destim~")
YIELD node, score
MATCH (node)-[:IS_ATTRIBUTE_OF]->(c:Compound)
WITH DISTINCT c as c, collect({{score: score, node: node}})[0] as s
WITH DISTINCT s as s, collect(c.pubChemCompId) as compoundId
RETURN s.node.name as name, s.node.pubChemSynId as synonymId, compoundId limit 2
Results:
{‘name’: ‘destim’,
‘synonymId’: ‘37d400b013a5db675a6fa80c629b8552’,
‘compoundId’: [‘compound:cid66124’, ‘compound:cid9306’]},
{‘name’: ‘bestim’,
‘synonymId’: ‘9c6c93d3b365a6768fb5a1f4285b7c1a’,
‘compoundId’: [‘compound:cid3038501’]}
Optimize Query
At the moment the query will match all nodes that have some match with the full-text search. This can become a lot if multiple words are involved or the matches afterwards do not add up to the limit of 5.
To reduce the time we first call the full-text search and limit the results to 50. This means the matching afterward will never have to deal with more than 50 nodes. Speeding up the query and reducing its memory usage.
CALL {
CALL db.index.fulltext.queryNodes("synonymsFullText", "hexahydro~ AND aminophenyl~ AND indeno~ AND pyridine~")
YIELD node, score
RETURN node, score limit 50}
MATCH (node)-[:IS_ATTRIBUTE_OF]->(c:Compound)
WITH DISTINCT c as c, collect({score: score, node: node})[0] as s
WITH DISTINCT s as s, collect(c.pubChemCompId) as compoundId
RETURN s.node.name as name, s.node.pubChemSynId as synonymId, compoundId limit 5
Splitting the Words
When calling the query above we need to find out what words we need to search for. For example, in “(+)-2,3,4,4a,5,9b-hexahydro-5-(4-aminophenyl)-1h-indeno(1,2-b)pyridine” everything which is not a combination of letters or numbers is discarded by Lucene full-text search, all letters that are not in the English alphabet will be replaced with English letters (öáì will become oai). This splitting is done in the backend with regex “[p{L}d]{2,}”. It will search for groups of Unicode characters (“p”) that are classified as letters (“{L}”) or a digit (“d”) with a length of 2 or more (“{2,}”). All of these will be followed by a “~” and joined with an “AND” so Lucene knows to fuzzy search for every word.
Because I remove all special characters it will also be more protected against Cypher injections.
Result front end
All of this together with a bit of front-end magic gives us the following application. We can see that “asprin” and “aspirine” are not detected at the same time, and spelling mistakes are fixed.

Full-text search in 197M chemical names graph database was originally published in Neo4j Developer Blog on Medium, where people are continuing the conversation by highlighting and responding to this story.