Kickstart a Chemical Graph Database

Back End Developer at CytoSMART
3 min read

I have spent some time scrapping and shaping PubChem data into a Neo4j graph database. The process took a lot of time, mainly downloading, and loading it into Neo4j. The whole process took weeks.
If you want to have your own I will show you how to download mine and set it up in less than an hour (most of the time you’ll just have to wait).

The process of how this dataset is created is described in the following blogs:
- Exploring Neodash for 197M chemical full-text graph
- Combining 3 Biochemical Datasets in a Graph Database
What Do You Get?
The full database is a merge of 3 datasets, PubChem (compounds + synonyms), NCI60 (GI50), and ChEMBL (cell lines).
It contains 6 nodes of interest:
Compound
This is related to a compound of PubChem. It has 1 property.
- pubChemCompId: The id within pubchem. So “compound:cid162366967” links to https://pubchem.ncbi.nlm.nih.gov/compound/162366967.
This number can be used with both PubChem RDF and PUG.
Synonym
A name found in the literature. This name can refer to zero, one, or more compounds. This helps find relations between natural language names and absolute compounds they are related to.
- Name: Natural language name. Can contain letters, spaces, numbers, and any other Unicode character.
- pubChemSynId: PubChem synonym id as used within the RDF
CellLine
These are the ChEMBL cell lines. They hold a lot of information.
- Name: The name of the cell line.
- Uri: A unique URI for every element within the ChEMBL RDF.
- cellosaurusId: The id to connect it to the Cellosaurus dataset. This is one of the most extensive cell line datasets out there.
Measurement
A measurement you can do within a biomedical experiment. Currently, only GI50 (the concentration needed for Growth Inhibition of 50%) is added.
- Name: Name of the measurement.
- Condition: A single condition of an experiment. A condition is part of an experiment. Examples are: an individual of the control group, a sample with drug A, or a sample with more CO2
Experiment
A collection of multiple conditions all done at the same time with the same bias. Meaning we assume all uncontrolled variables are the same.
- Name: Name of the experiment.

How Do Download It
Warning, you need 120 GB of free memory. The compressed file you download is already 30 GB. The uncompressed file is 30 GB. The database afterward is 60 GB.
60 GB is only for temporary files, the other 60 is for the database.
If you do this on an HDD hard disk it will be slow.
If you load this into Neo4j desktop as a local database (like I do) it will scream and yell at you, just ignore this. We are pushing it far further than it is designed for, but it will still work.
Download the File
Go to this Kaggle dataset and download the dump file. Unzip the file, then delete the zipped file. This part needs 60 GB but only takes 30 by the end of it.
Create a Database
Open the Neo4j desktop app, and click “Reveal files in File Explorer”. Move the `.dump` you downloaded into this folder.
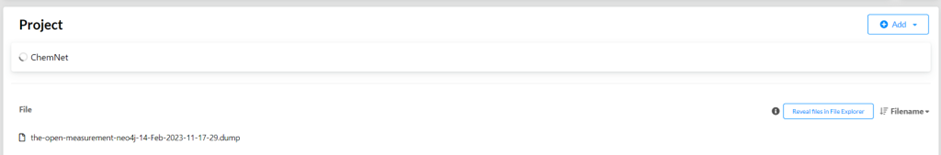
Click on the `…` behind the `.dump` file and click `Create new DBMS from dump`. This database is a dump from Neo4j V4, so your database also needs to be V4.x.x!

It will now create the database. This will take a long time, it might even say it has timed out. Do not believe this lie! In the background, it is still running.
Every time you start it, it will time out. Just let it run and press start later again. The second time it will be started up directly.
Every time I start it up I get the timed-out error. After waiting 10 minutes and clicking start again the database, and with it, more than 200 million nodes, is ready.
And you are done! Good luck and let me know what you build with it
Kickstart a chemical graph database was originally published in Neo4j Developer Blog on Medium, where people are continuing the conversation by highlighting and responding to this story.