Updating My Graph Database Using PubChems Endpoints

Back End Developer at CytoSMART
7 min read

In my last blog, I downloaded 197M nodes of all the chemical synonyms out of PubChem. This number was of course too low to cover every chemical synonym ever used.
I do want to connect the compounds from NCI (National Cancer Institute, USA) to the rest of the database. NCI gives every compound an NSC number (Cancer Chemotherapy National Service Center number), these are seen as synonyms by PubChem. Some of these NSC numbers are missing in the database, others are not connected to a compound.
Also, the data will get out of date, PubChem will update but my database will not. Besides that, some mistakes could have been introduced when loading the data. This means there is a need for a method to update synonyms and compounds in my local database.
PubChem Endpoints
Luckily PubChem has two great endpoints to work with. Less luckily, these two endpoints do not always give the same results. For this, we need to keep in mind how PubChem is structured. We are interested in three elements.
Synonym: one and only one name. This can be related to a compound and substance. Neither is limited. Compound: A standardized description. It can be related to multiple synonyms. Substance: A description found in a single record. It too can have multiple synonyms.
Because substances are seen as less scrutinized compounds, we only use the compounds. This does mean we have synonyms linking to nothing. This document explains the difference between substances and compounds better.
PUG (Power User Gateway)
PUG is a great tool to get direct information about a single object. If you look for a synonym name (like nsc815330), you will get the compound related to it together with the other known synonyms.
RDF (Resource Description Framework)
RDF (Resource Description Framework) is not focused on single objects, but on relationships. Searching by name is not possible, only by the id of the object. Searching for compound 46850181 gives us every object related to this compound and how they are related, from synonyms is-attribute-of this compound, to other compounds that are stereoisomers of this compound.
Difference
If we look for nsc815330 in PUG we get 4 synonyms: Melodinine K, CHEMBL1163260, NSC815330, and NSC-815330. All are part of compound 46850181. PUG does not allow me to search for synonyms based on the compound. Using RDF to search for the same compound 46850181 we find a link to 2 synonyms (Melodinine k, and CHEMBL1163260). Searching for the md5 encoded ID of NSC815330, NSC 815330, or NSC-815330 gives no results in RDF. This means both endpoints are different, not only in their use case but also in their data, and so should both be utilized.
Create Synonym ID
We can only search by id for synonyms in the RDF. But for that, we need to get the id from the name. A synonym ID is MD5_ + “MD5 hex hashing of the lowercase string of the name”. E.g. CHEMBL1163260 in lower case is chembl1163260, add MD5 hashing 388ea4f67de9a93d40f412a0fe85d044, add the prefix MD5_388ea4f67de9a93d40f412a0fe85d044 and we got the synonyms object in RDF found by id.
This was also used in my last blog, where I found that 5,865 of 197,000,000 synonyms did NOT follow this logic. This is close enough for our use case.
Backend Function
In order to use this logic more than once, I made a backend function out of it. This will be living in the same place as the search function.
Retrieving Data
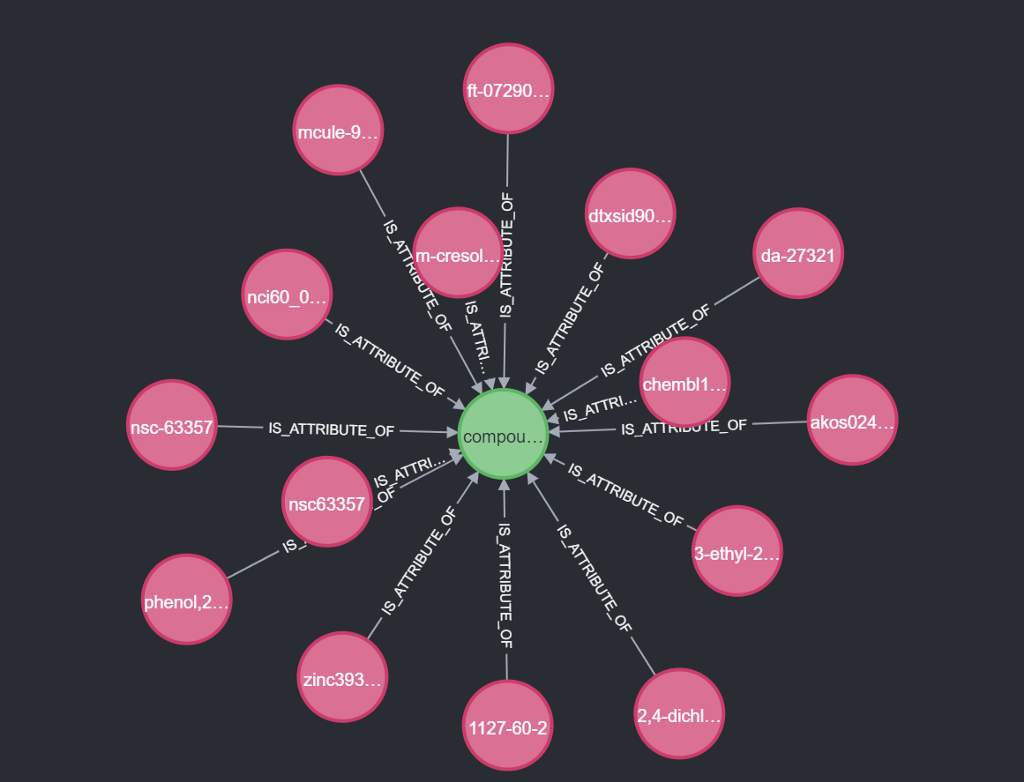
This function will check the PUG endpoint to find the compound and other synonyms, by name. For example “nsc63357” will give us this:
{
“CID”: 247901,
“Synonym”: [
“2,4-dichloro-3-ethyl-5-methylphenol”,
“1127–60–2”,
“Phenol,2,4-dichloro-3-ethyl-5-methyl-”,
“3-ethyl-2,4-dichloro-5-methyl-phenol”,
“NSC63357”,
“m-Cresol,6-dichloro-5-ethyl-”,
“CHEMBL1999840”,
“DTXSID90920869”,
“ZINC393690”,
“NSC-63357”,
“AKOS024332244”,
“MCULE-9983242334”,
“DA-27321”,
“NCI60_011195”,
“FT-0729068”
]
}
If none are found the function returns an empty list. This is very rare for NSC numbers. All the PUG results are given a constructed synonym ID (lowercase md5 hashing). The RDF is called with the compound ID. This gives us a list of synonym IDs connected to the compound (no names only IDs). Next up we combine these two lists and remove all duplicates based on ID. We give priority to PUG because RDF does not have names yet. If one of the synonyms does not have a name we make an HTTP call to the RDF synonym endpoint.
We are now left with a list of compounds. Each of these compounds has the given name (e.q. “nsc63357”) as a synonym. The compounds do hold all other synonyms too.
Updating the Database
For every compound, we will update the database. This will be done in two steps, removing wrong synonyms, and adding the right ones.
Removing incorrect synonyms.
The parts between curly brackets are python code or variables. We first find the compound by id and find all synonyms currently connected to it. Next up we filter them by the whole list of synonyms id we got from PUG and RDF calls. For everything not in the list, we remove the connection to this compound.
MATCH (c:Compound {{pubChemCompId: “{compound_id_str}”}})<-[r:IS_ATTRIBUTE_OF]-(s:Synonym)
WHERE NOT s.pubChemSynId IN {[i.id for i in synonyms]}
DELETE r
We only remove the connection because a synonym can be connected to other compounds.
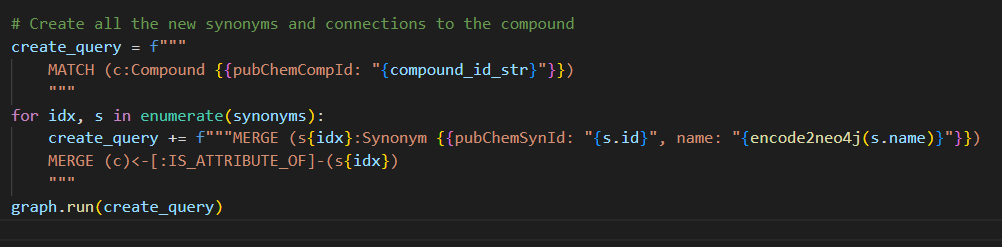
Adding correct synonyms
For this part, the python code does a bit more. Every synonym needs to be merged to prevent duplicates. We create a query with one match to get the compound. Followed by a line for every synonym to be merged. And then merge the relation between the synonym and compound. This creates a long query if there are a lot of synonyms but luckily python does that for us.
Updater in Action
For the next step, I want to add NCI60 data, in which every compound is given by its NSC number only. For a total of 56,685 unique NSC numbers in this dataset. But the synonym related to the NSC number 687296 can be nsc687296, nsc 687296, or nsc-687296 (capital letters do not matter for now). But there are also false positives like “nsc 687296/nsc 19893 combination”.
With Lucene, I searched for each NSC synonym, and if they have a compound, using the following query.
CALL {{
CALL db.index.fulltext.queryNodes(‘synonymsFullText’, “nsc687296 OR (nsc AND 687296)”)
YIELD node, score
return node limit 10
}}
OPTIONAL MATCH (node)-[:IS_ATTRIBUTE_OF]->(c:Compound)
RETURN node.name as name, node.pubChemSynId as synonymId, c.pubChemCompId as compoundId limit 5
This query has a very high recall but precision is lower, meaning it will find all synonyms we are looking for plus some more.
Next up we are going to filter the response, every synonym name that does not follow the nsc687296, nsc 687296, or nsc-687296 naming will be removed. Some of the synonym names we had to remove:
- n-(1-b-d-arabinofuranosyl-1,2-dihydro-2-oxo-4-pyrimidinyl)docosanamide, behenoylcytosine arabinoside, bh-ac, nsc-239336, sunrabin
- lentaron(r);17-dione;cgp-32349;nsc 282175
- nsc-d-611615
- nsc 625987, >=97% (HPLC)
- nsc 708781 incorporated in micelles, 0.55% (w/w) of drug
- nsc 708782 incorporated in micelles, 0.45% (w/w) of drug
- nsc 708783 incorporated in micelles, 0.20% (w/w) of drug
- nsc 708788 incorporated in micelles, 0.20% (w/w) of drug
- nsc-651016 free acid
This shows the 2 kinds of most common false positives we get, the nsc compound is used as part of a compound, and a list of synonyms that is not split into their own synonyms.
All of the nsc numbers also do have a true positive.
52847 of 56685 nsc numbers matched to at least one synonym with compound.
Update Synonyms With RDF
Some synonyms do NOT have a connection to a compound. We will make a call to the RDF endpoint to find out if we can find if this synonym has a known compound.
If it does we update the database with this new compound. There is a chance we need to create a new compound in the database. For that, we use MERGE.
8620 synonyms missed a compound, but after these RDF calls, only 3128 did. Note that every NSC number can have up to 3 synonyms.
These synonyms do not have to mean the NSC number was without a good synonym.
Use Full Update
For every NSC number without a good synonym, we make a call to the backend. We try 3 different synonyms before giving up, nsc123, nsc 123, and nsc-123. 2808 out of 3838 problematic NSC numbers are fixed this way.
Conclusion
If we now match all NSC numbers again we get 55578 out of 56685 NSC numbers matching at least one synonym with a compound. This means we can call this method a clear improvement of the current state.
Updating my graph database using PubChems endpoints was originally published in Neo4j Developer Blog on Medium, where people are continuing the conversation by highlighting and responding to this story.