Importing RDF Data
This chapter describes procedures that import RDF data into Neo4j.
The main method for importing RDF is n10s.rdf.import.fetch.
It imports and persists into Neo4j the triples returned by an URI.
This URI can point to an RDF file (local or remote) or a service producing RDF dynamically.
More on how to parameterise the access to web services in section Advanced settings for fetching RDF.
This section includes:
Procedure and Function Overview
The table below describes the available procedures and functions:
| type | qualified name | signature | description |
|---|---|---|---|
procedure |
|
|
Imports RDF from an url (file or http) and stores it in Neo4j as a property graph. Requires a unique constraint on :Resource(uri) |
procedure |
|
|
Imports an RDF snippet passed as parameter and stores it in Neo4j as a property graph. Requires a unique constraint on :Resource(uri) |
function |
|
|
Returns the expanded (full) IRI given a shortened one created in the load process with semantics.importRDF |
function |
|
|
Returns the XMLSchema or custom datatype of a property when present |
function |
|
|
Returns the local part of an IRI |
function |
|
|
Returns the namespace part of an IRI |
function |
|
|
Returns the language tag of a value. Returns null if the value is not a string orif the string has no language tag |
function |
|
|
Returns the first value with the language tag passed as first argument or null if there’s not a value for the provided tag |
function |
|
|
Returns the value of a datatype of a property after stripping out the datatype information when present |
function |
|
|
Returns false if the value is not a string or if the string is not tagged with the given language tag |
function |
|
|
Returns the shortened version of an IRI using the existing namespace definitions |
Parameters
The import procedures take the following three parameters:
| Parameter | Type | Description |
|---|---|---|
url |
String |
URL of the dataset |
format |
String |
serialization format. Valid formats are: Turtle, N-Triples, JSON-LD, RDF/XML, TriG and N-Quads (For named graphs) |
params |
Map |
Optional set of parameters (see description in table below) |
| If we try to run any RDF import procedure without having defined the uniqueness constraint or created a GraphConfig first, the import will fail showing an error message indicating what’s missing. |
The RDF import procedures
Once a basic GraphConfig has been created, we can call the RDF import procedures.
Let’s look at the first of them:
In its most basic form the n10s.rdf.import.fetch method takes a URL string to access the RDF data and the serialisation format.
Let’s say you’re trying to load the following RDF document into Neo4j.
@prefix neo4voc: <http://neo4j.org/vocab/sw#> .
@prefix neo4ind: <http://neo4j.org/ind#> .
neo4ind:nsmntx3502 neo4voc:name "NSMNTX" ;
a neo4voc:Neo4jPlugin ;
neo4voc:version "3.5.0.2" ;
neo4voc:releaseDate "03-06-2019" ;
neo4voc:runsOn neo4ind:neo4j355 .
neo4ind:apoc3502 neo4voc:name "APOC" ;
a neo4voc:Neo4jPlugin ;
neo4voc:version "3.5.0.4" ;
neo4voc:releaseDate "05-31-2019" ;
neo4voc:runsOn neo4ind:neo4j355 .
neo4ind:graphql3502 neo4voc:name "Neo4j-GraphQL" ;
a neo4voc:Neo4jPlugin ;
neo4voc:version "3.5.0.3" ;
neo4voc:releaseDate "05-05-2019" ;
neo4voc:runsOn neo4ind:neo4j355 .
neo4ind:neo4j355 neo4voc:name "neo4j" ;
a neo4voc:GraphPlatform , neo4voc:AwesomePlatform ;
neo4voc:version "3.5.5" .You can save it to your local drive or access them directly here.
All you’ll need to provide to n10s is the location (file:// or http://) and the serialisation used, Turtle in this case.
If you’re working on a windows machine and want to access an RDF file stored on your drive, here’s the syntax for paths: CALL n10s.rdf.import.fetch("file:///D:\\Data\\some_rdf_file.rdf", "RDF/XML");
|
The following imports the nsmntx.ttl file hosted on GitHub
CALL n10s.rdf.import.fetch("https://github.com/neo4j-labs/neosemantics/raw/3.5/docs/rdf/nsmntx.ttl","Turtle");n10s will import the RDF data and persist it into your Neo4j graph as the following structure
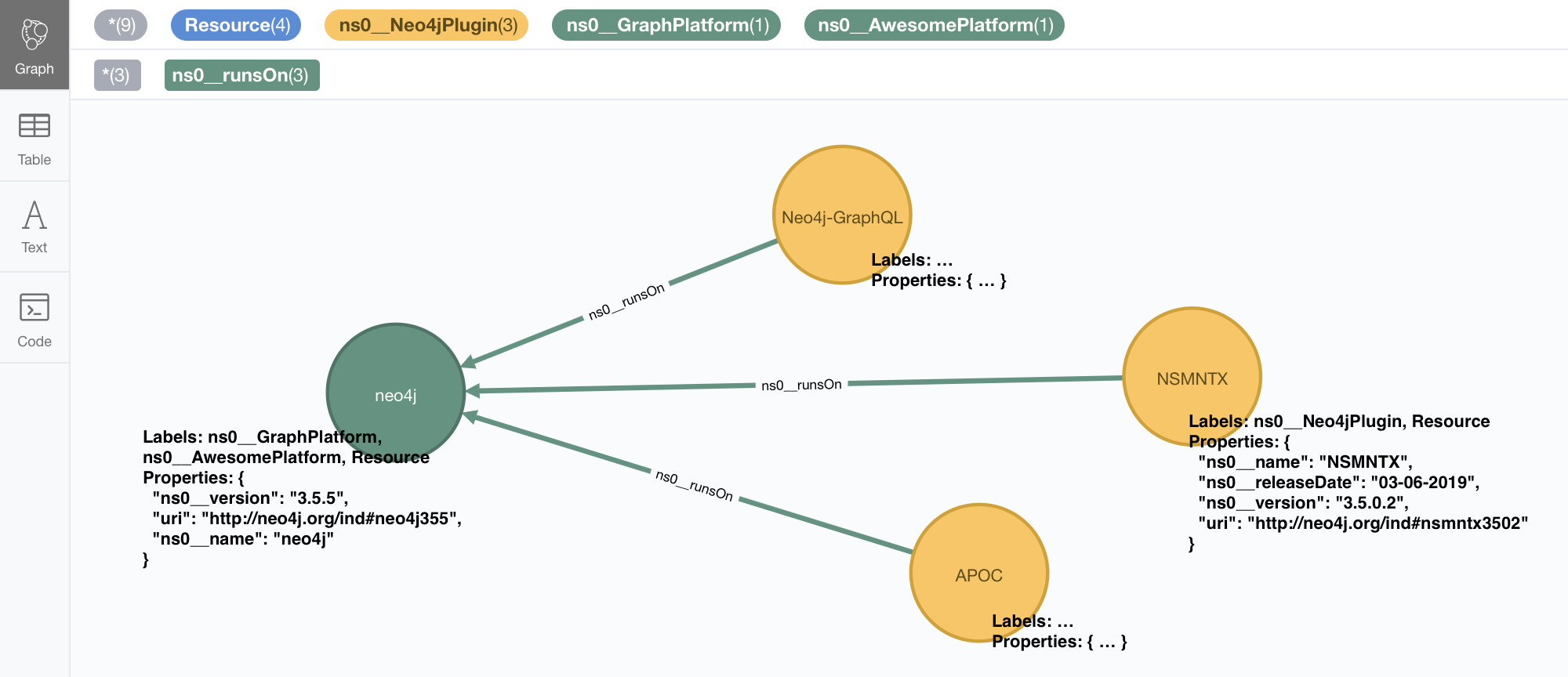
The first thing we notice is that dataType properties in your RDF have been converted into node properties and object properties are now relationships connecting nodes. Every node represents a resource and has a property with its uri. Similarly, rdf:type statements are transformed into node labels. That’s pretty much it but if you are interested, there is a complete description of the way triple data is transformed into Property Graph data for storage in Neo4j in this post. You will also notice a terminology/vocabulary transformation applied by default. The URIs identifying the elments in the RDF data (resources, properties, etc) have their namespace part shortened to make them more human readable and easier to query with Cypher.
In our example, http://neo4j.org/vocab/sw#name has been shortened to ns0_\_name (notice the double underscore separator used between the prefix and the local name in the URI).
Similarly, http://www.w3.org/1999/02/22-rdf-syntax-ns#type would be shortened to rdf\_\_type and so on…
Prefixes for custom namespaces are assigned dynamically in sequence (ns0, ns1, etc) as they appear in the imported RDF.
This is the default behavior but we’ll see later on that it is possible to control that, and use custom prefixes.
More details in section Defining custom prefixes for namespaces.
Keeping namespaces can be important if you care about being able to regenerate the imported RDF as we will see in section Exporting RDF data.
However, if you don’t care about that, you can tell Neo4j to ignore the namespaces by setting
the handleVocabUris parameter to 'IGNORE' in the GraphConfig.
CALL n10s.graphconfig.set({ handleVocabUris: "IGNORE" });After applying this setting, namespaces will be discarded on all subsequent imports.
So if you run the import, only the local names of URIs will be kept.
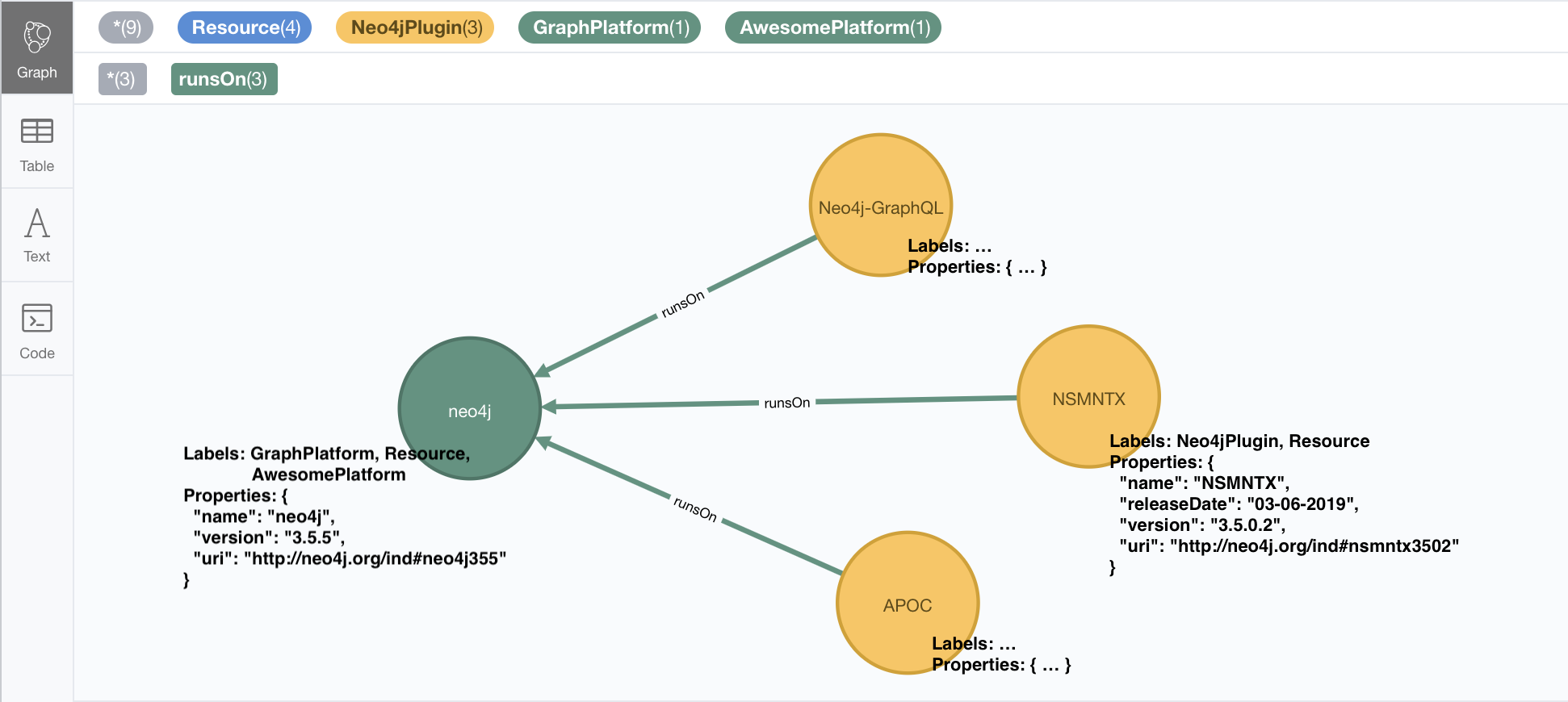
The importRDF method call is identical: here’s what that would look like:
CALL n10s.rdf.import.fetch("https://github.com/neo4j-labs/neosemantics/raw/3.5/docs/rdf/nsmntx.ttl","Turtle");In this case, the imported graph will look something like this, in which the names for labels, properties and relationships are more of the kind you’re use to work with in Neo4j:
| The first great thing about getting your RDF data into Neo4j is that now you can query it with Cypher |
Here’s an example that showcases the difference: Let’s say you want to produce a list of plugins that run on Neo4j and what’s the latest versions of each.
If your RDF data is stored in a triple store, you would need to use the SPARQL query on the left to answer the question. To the right you can see the same thing expressed with Cypher in Neo4j.
| SPARQL | Cypher |
|---|---|
|
|
We’ve seen how to shorten RDF uris into more readable names using namespace prefixes, and we’ve also seen how to ignore them completely.
There is a third option: You can keep the complete uris in property names, labels and relationships
in the graph by setting the handleVocabUris property to "KEEP". The result will not be pretty
and your cypher queries on the imported model will look pretty horrible, but hey, the option is there.
The following deletes existing Resource nodes and updates the GraphConfig
// n10s created nodes must be deleted before updating Graph Config
MATCH (resource:Resource) DETACH DELETE resource;
CALL n10s.graphconfig.set({ handleVocabUris: "KEEP" })And we can re-import with the usual command:
CALL n10s.rdf.import.fetch("https://github.com/neo4j-labs/neosemantics/raw/3.5/docs/rdf/nsmntx.ttl","Turtle"})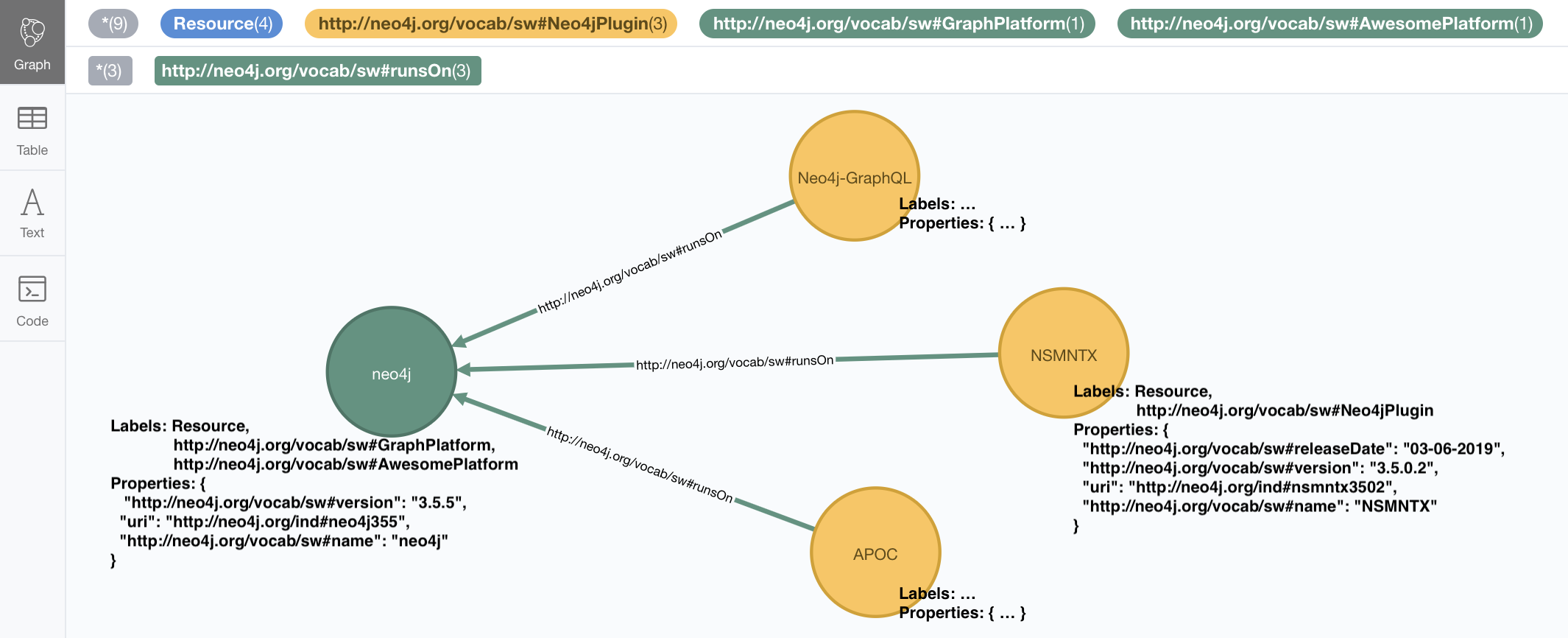
The imported graph in this case has the same structure, but uses full URIs as labels, relationships, and property names.
Importing RDF passed as parameter
So far we have always assumed the RDF to be imported is retrived from a URL (whether a static file in the previous
examples or dynamically generated as the result of a SPARQL query as described in Advanced settings for fetching RDF).
The reality is that sometimes the RDF is not retrieved from a URL but passed as a paramter to the import procedure.
For this type of use Neosemantics offers a set of procedures named *.inline instead of *.fetch.
Here is an example:
WITH '
<neo4j://individual/JB> <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://neo4j.org/voc#Person> .
<neo4j://individual/JB> <http://neo4j.org/voc#name> "J. Barrasa" .
<neo4j://individual/JB> <http://neo4j.org/voc#altName> "Dr J" .
' as payload
CALL n10s.rdf.import.inline(payload,"N-Triples") YIELD terminationStatus, triplesLoaded
RETURN terminationStatus, triplesLoadedSimilarly the RDF fragment can be defined as a parameter (here’s an example of how to do it in the browser):
:PARAM payload:
<neo4j://individual/JB> <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://neo4j.org/voc#Person> .
<neo4j://individual/JB> <http://neo4j.org/voc#name> "J. Barrasa" .
<neo4j://individual/JB> <http://neo4j.org/voc#altName> "Dr J" .And then used in the import request:
CALL n10s.rdf.import.inline($payload,"N-Triples")
For every import procedure there is a .fetch and a .inline version.
|
Filtering triples by predicate
Something you may need to do when importing RDF data into Neo4j is exclude certain triples so that they are not persisted in your Neo4j graph.
This is useful when only a portion of the RDF data is relevant to your project.
The exclusion is done by predicate type "I don’t need to load the version property, or the release date", all you’ll
need to do is provide the list of URIs of the predicates you want excluded in parameter predicateExclList.
Note that the list needs to contain full URIs.
The following imports data from the nsmntx.ttl Turtle file excluding triples with version or releaseDate as predicates:
CALL n10s.rdf.import.fetch("https://github.com/neo4j-labs/neosemantics/raw/3.5/docs/rdf/nsmntx.ttl", "Turtle", {
predicateExclusionList : [ "http://neo4j.org/vocab/sw#version", "http://neo4j.org/vocab/sw#releaseDate" ]
});Handling multivalued properties
In RDF multiple values for the same property are just multiple triples. For example, you can have multiple alternative names for an individual like in the next RDF fragment:
<neo4j://individual/JB> <http://www.w3.org/1999/02/22-rdf-syntax-ns#type> <http://neo4j.org/voc#Person> .
<neo4j://individual/JB> <http://neo4j.org/voc#name> "J. Barrasa" .
<neo4j://individual/JB> <http://neo4j.org/voc#altName> "JB" .
<neo4j://individual/JB> <http://neo4j.org/voc#altName> "Jesús" .
<neo4j://individual/JB> <http://neo4j.org/voc#altName> "Dr J" .n10s default behavior is to keep only one value for literal properties and it will be the last one read in the triples parsed. So if you run a straight import on that data like this:
The following deletes existing Resource nodes and updates the GraphConfig
// n10s created nodes must be deleted before updating Graph Config
MATCH (resource:Resource) DETACH DELETE resource;
CALL n10s.graphconfig.init();The following imports data from the multivalued1.nt N-Triples file
CALL n10s.rdf.import.fetch("https://github.com/neo4j-labs/neosemantics/raw/3.5/docs/rdf/multivalued1.nt","N-Triples");Only the last value for the multivalued altName property will be kept.
MATCH (n:ns0__Person)
RETURN n.ns0__name as name, n.ns0__altName as altName;| name | altName |
|---|---|
"J. Barrasa" |
"Dr J" |
This makes things simple and will be perfect if your dataset does not have multivalued properties.
It can also be fine if keeping only one value is acceptable, either because the property is not critical or because one value is enough.
There will be other cases though, where we do need to keep all the values, and here’s where the config parameter handleMultival will help.
Here’s how:
The following deletes existing Resource nodes and updates the GraphConfig
// n10s created nodes must be deleted before updating Graph Config
MATCH (resource:Resource) DETACH DELETE resource;
CALL n10s.graphconfig.set({ handleMultival: 'ARRAY' });The following imports data from the multivalued1.nt N-Triples file
CALL n10s.rdf.import.fetch("https://github.com/neo4j-labs/neosemantics/raw/3.5/docs/rdf/multivalued1.nt","N-Triples");Now all properties are stored as arrays in Neo4j. Even the ones that have one value only! But we can do better than that, let’s have a look at another example.
The following Turtle RDF fragment with the description of a news article. The article has a number of keywords associated with it.
@prefix og: <http://ogp.me/ns#> .
@prefix nyt: <http://nyt.com/voc/> .
<nyt://article/a17a9514-73e7-51be-8ade-283e84a6cd87>
a og:article ;
og:title "Bengal Tigers May Not Survive Climate Change" ;
og:url "https://www.nytimes.com/2019/05/06/science/tigers-climate-change-sundarbans.html" ;
og:description "The tigers of the Sundarbans may be gone in fifty years, according to study" ;
nyt:keyword "Climate Change", "Endangered Species", "Global Warming", "India", "Poaching" .We want to make sure we keep all values for the nyt:keyword property. The natural way to
do this in Neo4j is storing them in an array, so we’ll instruct Neosemantics to do that by setting
in the GraphConfig the parameters handleMultival to 'ARRAY' and multivalPropList to the list
of property types that are multivalued and we want stored as arrays of values.
In the previous example the list will only contain 'http://nyt.com/voc/keyword'.
Here’s the config setting and import commands that we need.
Note that I’m combining the multivalued property config setting with the handleVocabUris set
to false (the interested reader can try to drop this config and get URIS shortened with prefixes instead):
The following deletes existing Resource nodes and updates the GraphConfig
// n10s created nodes must be deleted before updating Graph Config
MATCH (resource:Resource) DETACH DELETE resource;
CALL n10s.graphconfig.set({
handleVocabUris: "IGNORE",
handleMultival: 'ARRAY',
multivalPropList : ['http://nyt.com/voc/keyword']
});The following imports data from the multivalued2.ttl Turtle file
CALL n10s.rdf.import.fetch("https://github.com/neo4j-labs/neosemantics/raw/3.5/docs/rdf/multivalued2.ttl","Turtle");And here’s what the result of the import would look like:
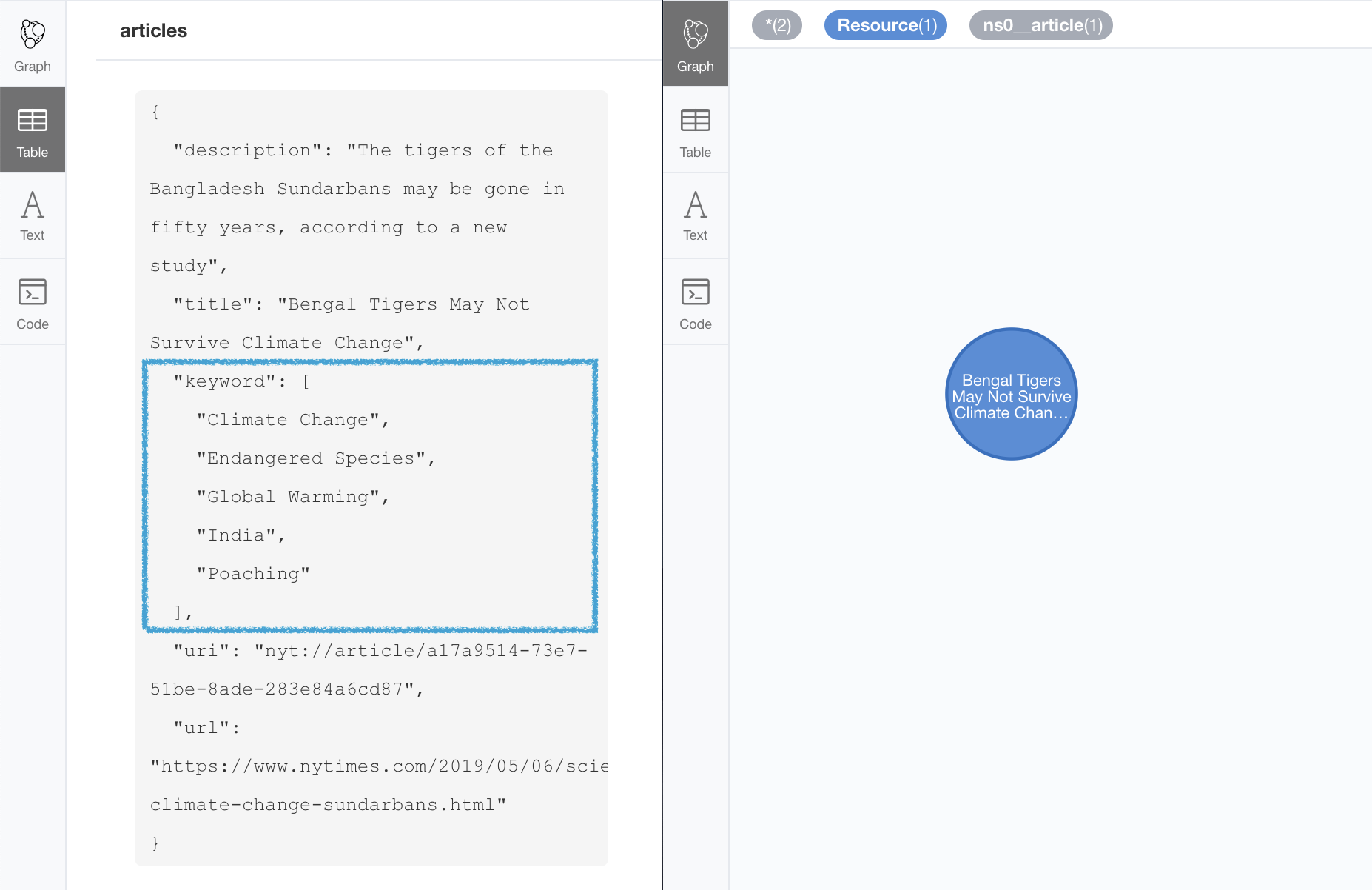
When we analyse the result in the Neo4j browser we realise that there’s only one node for the nine triples imported! Yes, keep in mind that all triples in our RDF fragment are datatype properties, or in other words, properties with literal values, which are stored in Neo4j as node properties. All the statements are there, no data is lost, it’s just stored as the internal structure of the node. We can see all properties on the table view on the left hand side of the image.
Note that this time only the properties enumerated in the multivalPropList config parameter are stored as arrays, the rest are kept as atomic values.
Remember that if we set handleMultival to 'ARRAY' in our GraphConfig but we don’t provide a
list of property URIs as multivalPropList ALL literal properties will be stored as arrays in every subsequent import.
|
Here’s an example of how to query the multiple values of the keyword property: Give me articles tagged with the "Global Warming" keyword.
MATCH (a:article)
WHERE "Global Warming" IN a.keyword
RETURN a.title as title;| title |
|---|
"Bengal Tigers May Not Survive Climate Change" |
Handling language tags
Literal values in RDF can be tagged with language information. This can be used in any context
but it’s common to find it used in combination with multivalued properties to create multilingual
descriptions for items in a dataset. In the following example we have a description of a TV
series with a multivalued property show:localName where each of the values is annotated with the
language.
@prefix show: <http://example.org/vocab/show/> .
@prefix indiv: <http://example.org/ind/> .
ind:218 a show:TVSeries
ind:218 show:name "That Seventies Show" .
ind:218 show:localName "That Seventies Show"@en .
ind:218 show:localName 'Cette Série des Années Soixante-dix'@fr .
ind:218 show:localName "Cette Série des Années Septante"@fr-be .By default, Neosemantics will strip out the language tags but if you want to keep them you’ll need to
set the keepLangTag to true. If we uset it in combination with the setting required to keep
all values of a property stored in an array, the import invocation would look something like this:
The following deletes existing Resource nodes and updates the GraphConfig
// n10s created nodes must be deleted before updating Graph Config
MATCH (resource:Resource) DETACH DELETE resource;
CALL n10s.graphconfig.set({
keepLangTag: true,
handleMultival: 'ARRAY',
multivalPropList : ['http://example.org/vocab/show/localName']
});The following imports data from the multilang.ttl Turtle file
CALL n10s.rdf.import.fetch("https://github.com/neo4j-labs/neosemantics/raw/3.5/docs/rdf/multilang.ttl","Turtle");When you import literal values keeping the language annotation, you’ll see that string values have a suffix like @fr for French language, @zh-cmn-Hant for Mandarin Chinese traditional, and so on.
The function getLangValue can be used to get the value for a particular language tag. It returns null when there is not a value for the selected language tag.
The following Cypher fragment returns the french version of a property and when not found, defaults to the english version.
MATCH (n:Resource)
RETURN coalesce(n10s.rdf.getLangValue("fr",n.ns0__localName), n10s.rdf.getLangValue("en",n.ns0__localName));There are two additional functions for handling language-tagged property values: n10s.rdf.hasLangTag and n10s.rdf.getLangTag.
-
n10s.rdf.getLangTagreturns the language tag (when present) from a property value. -
n10s.rdf.hasLangTagreturns true if a property value is tagged with a given language tag or false if not.
Here is an example of both functions:
First we create a node with a multivalued property where each value is language-tagged:
CREATE (n:Thing {
prop: [
"That Seventies Show@en-US",
"Cette Série des Années Soixante-dix@fr-custom-tag",
"你好@zh-Hans-CN"
]
});We can use n10s.rdf.getLangTag to get the language tag for any specific value
MATCH (n:Thing)
RETURN n10s.rdf.getLangTag(n.prop[0]) AS tag;| tag |
|---|
"en-US" |
or for all of them:
MATCH (n:Thing)
RETURN [x in n.prop | n10s.rdf.getLangTag(x)] AS tags;| tags |
|---|
["en-US", "fr-custom-tag", "zh-Hans-CN"] |
And similarly, we can use n10s.rdf.hasLangTag to check whether the first value matches a given language tag:
MATCH (n:Thing)
RETURN n10s.rdf.getValue(n.prop[0]) as val,
n10s.rdf.hasLangTag("es",n.prop[0]) as isSpanish;| val | isSpanish |
|---|---|
"That Seventies Show" |
FALSE |
Filtering triples by language tag
Language tags can also be used as a filter criteria. If we are only interested in a particular
language when loading a multilingual dataset, we can set a filter so only literal values with a
given language tag (or untagged ones) are imported into Neo4j. The configuration parameter that
does it is languageFilter and you’ll need to set it to the relevant tag, for instance 'es' for
literals in Spanish language.
Note that this config is request specific, as we may want to import certain language tags from
a file, and others from a different one. That’s why instead of being in the GraphConfig it’s in
the import method param section. Here’s what it would look like:
CALL n10s.rdf.import.fetch("https://github.com/neo4j-labs/neosemantics/raw/3.5/docs/rdf/multilang.ttl","Turtle", {
languageFilter: 'es'
});Handling custom data types
In RDF custom data types are annotated to literals after the seperator ^^ in form of an IRI.
For example, you can have a custom data type for a currency like in the following Turtle RDF fragment:
@prefix ex: <http://example.com/> .
ex:Mercedes
rdf:type ex:Car ;
ex:price "10000"^^ex:EUR ;
ex:power "300"^^ex:HP ;
ex:color "red"^^ex:Color .The default behavior of neosemantics is to not keep custom data types for properties. So if you run a straight import on that data like this:
CALL n10s.rdf.import.fetch("file://tmp/customDataTypes.ttl","Turtle")Only the value for the properties will be kept.
MATCH (n:ns0__Car)
RETURN n.ns0__price, n.ns0__power, n.ns0__color;| n.ns0__price | n.ns0__power | n.ns0__color |
|---|---|---|
"10000" |
"300" |
"red" |
This makes things simple and will be perfect if your dataset does not have properties with custom data types.
But if you need to keep the custom data types, you’ll want to set the config parameter keepCustomDataTypes in your GraphConfig.
Here’s how:
The following deletes existing Resource nodes and updates the GraphConfig
// n10s created nodes must be deleted before updating Graph Config
MATCH (resource:Resource) DETACH DELETE resource;
CALL n10s.graphconfig.set({keepCustomDataTypes: true});The following imports data from the customDataTypes.ttl Turtle file
CALL n10s.rdf.import.fetch("https://github.com/neo4j-labs/neosemantics/raw/3.5/docs/rdf/customDataTypes.ttl","Turtle");Now all properties that have a custom data type are saved as strings with their respective custom data type IRIs in Neo4j.
| n.ns0__price | n.ns0__power | n.ns0__color |
|---|---|---|
"10000^^ns0__EUR" |
"300^^ns0__HP" |
"red^^ns0__Color" |
But we can do better than that, let’s have a look at another example. We will use the same Turtle file from above for this example.
If we want to keep the custom data type for only some of the properties then we can instruct neosemantics
to do that by setting keepCustomDataTypes to true and customDataTypePropList to the list of property
types whose custom data types we want to keep.
In the example the list will only contain 'http://example.com/power'.
Here is the import command that we need:
// n10s created nodes must be deleted before updating Graph Config
MATCH (resource:Resource) DETACH DELETE resource;
CALL n10s.graphconfig.set({
keepCustomDataTypes: true,
customDataTypePropList: ['http://example.com/power']
});
CALL n10s.rdf.import.fetch("https://github.com/neo4j-labs/neosemantics/raw/3.5/docs/rdf/customDataTypes.ttl","Turtle");And here’s what the result of the cypher query above would look like after this import:
| n.ns0__price | n.ns0__power | n.ns0__color |
|---|---|---|
"10000" |
"300^^ns0__HP" |
"red" |
Note that this time only the custom data types of the properties listed in the customDataTypedPropList are kept, the rest will only have the literal value.
Remember that if we set keepCustomDataTypes to true but we don’t provide a list of property URIs as customDataTypedPropList ALL literals with a custom data type will be stored as strings with their respective custom data type IRIs.
|
When you import literal values keeping the custom data types, you’ll see that string values have a IRI suffix separated by ^^ from the raw value. For instance "10000^^ns0__EUR" from the example above.
The function getDataType can be used to get the data type for a particular property. It returns null when there is no custom data type for the given property.
The following Cypher fragment returns the data type of power.
MATCH (n:ns0__Car)
RETURN n10s.rdf.getDataType(n.ns0__power);The function getValue can be used to get the raw value of a particular property without custom data types or language tags.
The following Cypher fragment returns the raw value of power.
MATCH (n:ns0__Car)
RETURN n10s.rdf.getValue(n.ns0__power);The user functions mentioned above can be combined with other user functions like uriFromShort or getIRILocalName etc.
Classes as Nodes (instead of Labels)
The rdf:type statements in RDF (triples) are transformed into labels by default when we import them into Neo4j. While this is a reasonable approach it may not be your preferred option, especially if you want to load an ontology too and link it to your instance data. In that case you’ll probably want to represent the types as nodes and have 'the magic' of uris have them linked.
Note that this approach can create very dense nodes when loading large datasets.
By setting the handleRDFTypes parameter in the graph config to "LABELS_AND_NODES" we can achieve exactly that behavior.
Actually, as the name suggests we get a double effect and rdf:type statements (triples)
are persisted both as labels and as rdf_type relationships connecting the individual to a node
representing the class.
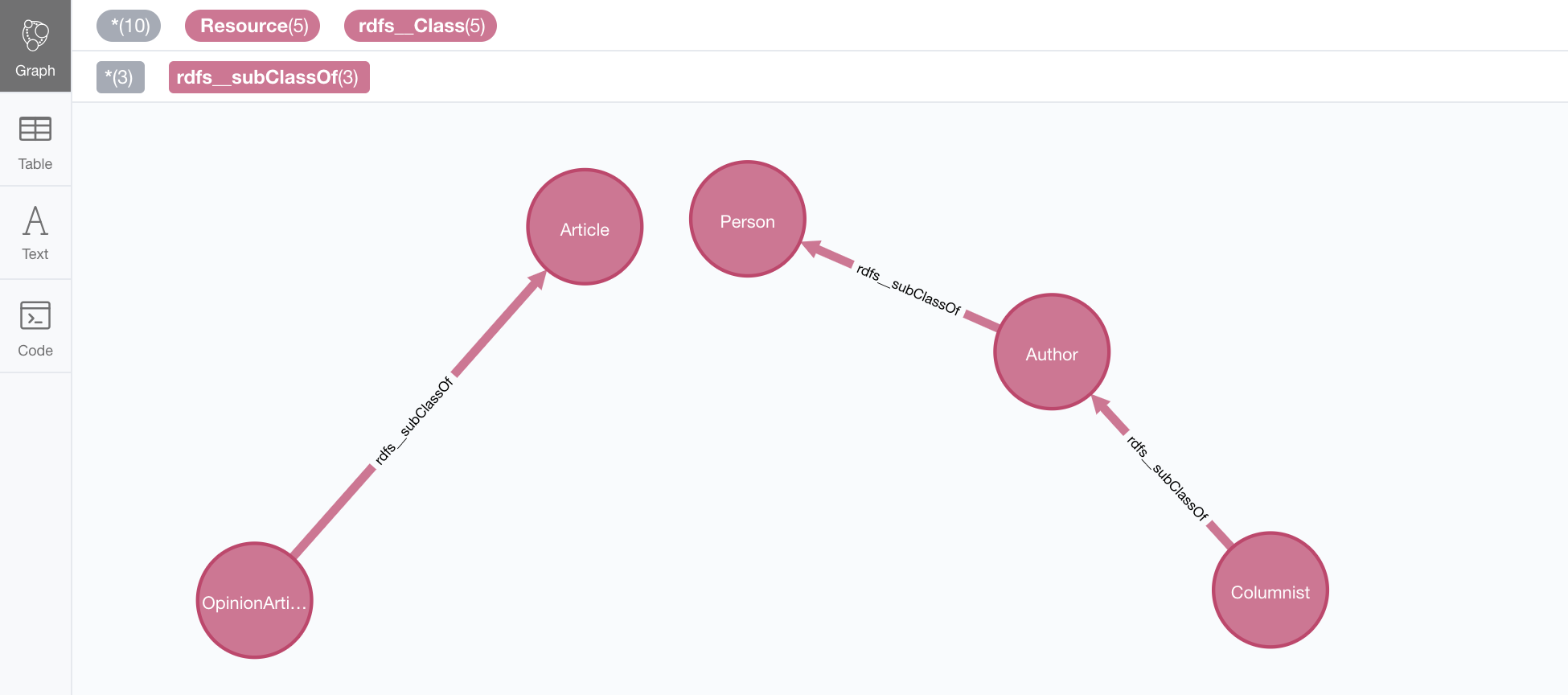
Here’s an example: Let’s say we want to load an ontology (notice that it’s actually a small fragment of several ontologies, but it will work for our example). For what it’s worth, it’s an RDF file, so we load it the usual way, with all default settings
CALL n10s.graphconfig.init({handleRDFTypes:"LABELS_AND_NODES"});
CALL n10s.rdf.import.fetch("https://github.com/neo4j-labs/neosemantics/raw/3.5/docs/rdf/minionto.ttl","Turtle");We can inspect the result of the import to see that the ontology contains the five class definitions
linked in a hierarchy as in the following image.
(there are additional rdf_type relationships not shown in this image, linking each class node to an rdfs:Class node).
Next we want to import the instance data, and we want it to link to the ontology graph rather than build a
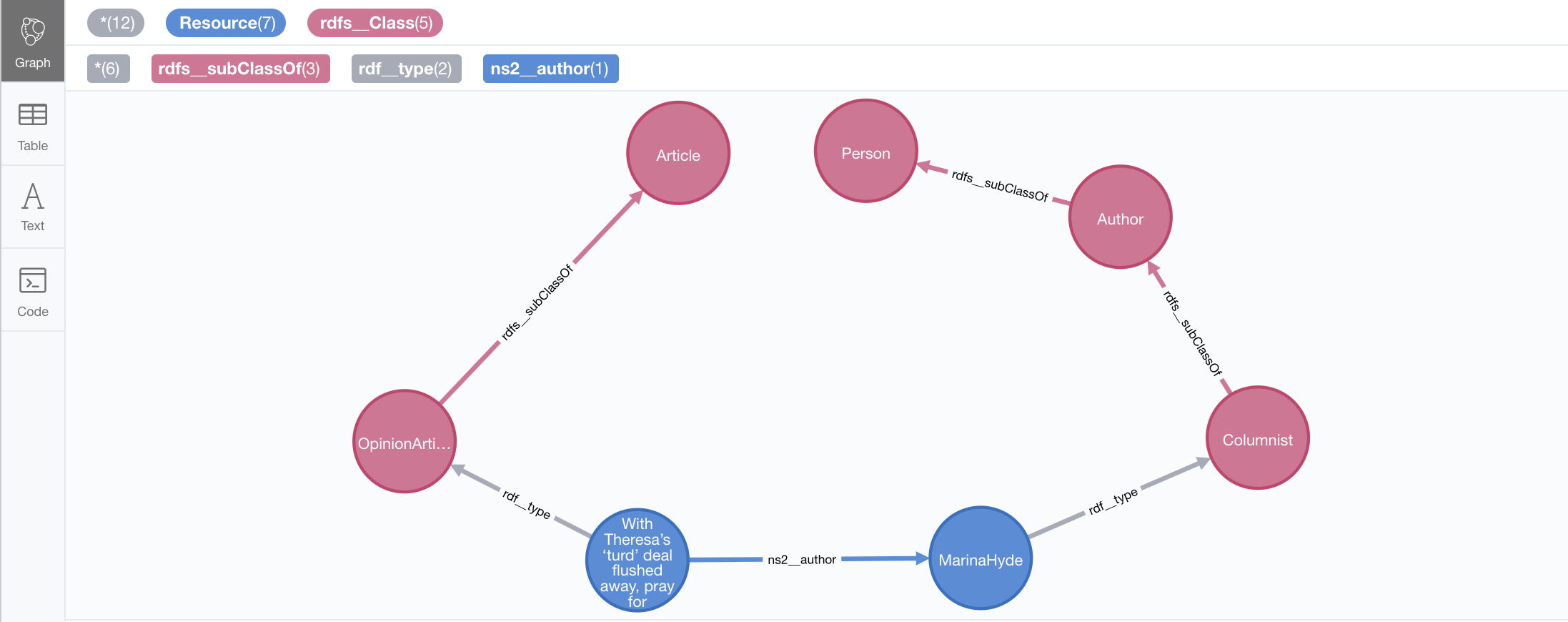
disconnected graph (this is what would happen if rdf:type statements were imported only as labels).
With the current "LABELS_AND_NODES" setting, we get the types as label but also as relationships connecting
the instances to the previously loaded ontology.
call n10s.rdf.import.fetch("https://github.com/neo4j-labs/neosemantics/raw/3.5/docs/rdf/miniinstances.ttl","Turtle");The resulting graph connects the instance data to the ontology elements. This is the magic of unique identifiers (uris), tere’s nothing you need to do for the linkage to happen, if your RDF is well formed and uris are used consistently in it, then it will happen automatically.
There is a third option for the handleRDFTypes configuration parameter, it is "NODES" and as its name
suggests persists rdf:type statements exclusively as relationships.
More on the usefulness of representing the ontology in the neo4j graph in section Inference/Reasoning.
Advanced settings for fetching RDF
Sometimes the RDF data will be a static file, and other times it’ll be dynamically generated in response to an HTTP request (GET or POST) possibly containg parameters, even a SPARQL query.
The following two parameters will help in these situations:
payload : Takes a String as value and sends the specified data in a POST HTTP request to the the url passed as first parameter of the Stored Procedure. Useful typicaloy for SPARQL endpoints where we want to submit a query to produce the actual RDF.
headerParams : Takes a map of property-values and adds each of them as an extra header in the HTTP request. Useful for sending credentials to services requiring authentication (using Authorization header) or to specify the required format (using Accept header).
Here is an example of how to send a request to a SPARQL endpoint and ingest the results directly in Neo4j. The service in question is the Linked Open Data service of the British Library. You can test it here. The service is not authenticated, so no need to use the Authorization header but we want to select the RDF serialisation produced by our request, which we do by setting Accept: "application/turtle". Finally, we pass the SPARQL query as the value of the payload parameter, prefixed with query=.
headerParams: { Accept: "application/turtle"}, payload: "query=DESCRIBE <http://bnb.data.bl.uk/id/resource/018212405>" }
We obviously need a query producing RDF so we can import it into Neo4j. I’m using a SPARQL DESCRIBE query in the following example but a SPARQL CONSTRUCT query could be used too. If you want to import all the details available in the British Library about 'The world of yesterday' by Stefan Zweig’s, which by the way, if you haven’t read, you should really take a break after this section and go read.
The following deletes all Resource nodes and sets config to ignore vocab URIs
// n10s created nodes must be deleted before updating Graph Config
MATCH (resource:Resource) DETACH DELETE resource;
CALL n10s.graphconfig.set({handleVocabUris: "IGNORE"});-
The following imports data from the British Library’s SPARQL API
CALL n10s.rdf.import.fetch("https://bnb.data.bl.uk/sparql","Turtle", {
handleVocabUris: "IGNORE",
headerParams: { Accept: "application/turtle"},
payload: "query=" + apoc.text.urlencode("DESCRIBE <http://bnb.data.bl.uk/id/resource/018212405>")
});
Notice that the British Library service requires you to encode the SPARQL query. We do this with APOC’s apoc.text.urlencode function.
After running this you get a pretty poor graph, because the DESCRIBE query only returns the statements having 'The world of yesterday' (http://bnb.data.bl.uk/id/resource/018212405) as subject or object.
But we can enrich it a bit by re-running it for a all of the URIs connected to our book as follows:
MATCH (:Book)-->(t) WITH DISTINCT t
CALL n10s.rdf.import.fetch("https://bnb.data.bl.uk/sparql","Turtle", {
handleVocabUris: "IGNORE",
headerParams: { Accept: "application/turtle"},
payload: "query=" + apoc.text.urlencode("CONSTRUCT {<" + t.uri + "> ?p ?o } { <" + t.uri + "> ?p ?o } LIMIT 10 ")
})
yield triplesLoaded
return t.uri, triplesLoaded;Which returns:
| t.uri | triplesLoaded |
|---|---|
"http://bnb.data.bl.uk/id/place/Europe" |
3 |
"http://bnb.data.bl.uk/id/resource/GBB721847" |
1 |
"http://rdaregistry.info/termList/RDAContentType/1020" |
1 |
"http://lexvo.org/id/iso639-3/eng" |
0 |
"http://bnb.data.bl.uk/id/concept/ddc/e22/838.91209" |
3 |
"http://bnb.data.bl.uk/id/person/ZweigStefan1881-1942" |
5 |
"http://bnb.data.bl.uk/id/concept/place/lcsh/Europe" |
4 |
"http://bnb.data.bl.uk/id/concept/lcsh/WorldWar1914-1918Influence" |
5 |
"http://rdaregistry.info/termList/RDAMediaType/1003" |
1 |
"http://rdaregistry.info/termList/RDACarrierType/1018" |
1 |
"http://bnb.data.bl.uk/id/concept/person/lcsh/ZweigStefan1881-1942" |
5 |
"http://bnb.data.bl.uk/id/concept/lcsh/AuthorsAustrian20thcenturyBiography" |
5 |
"http://bnb.data.bl.uk/id/concept/lcsh/EuropeCivilization20thcentury" |
5 |
"http://bnb.data.bl.uk/id/resource/018212405/publicationevent/PlaceofpublicationnotidentifiedPushkinPress2009" |
4 |
And produces this graph:
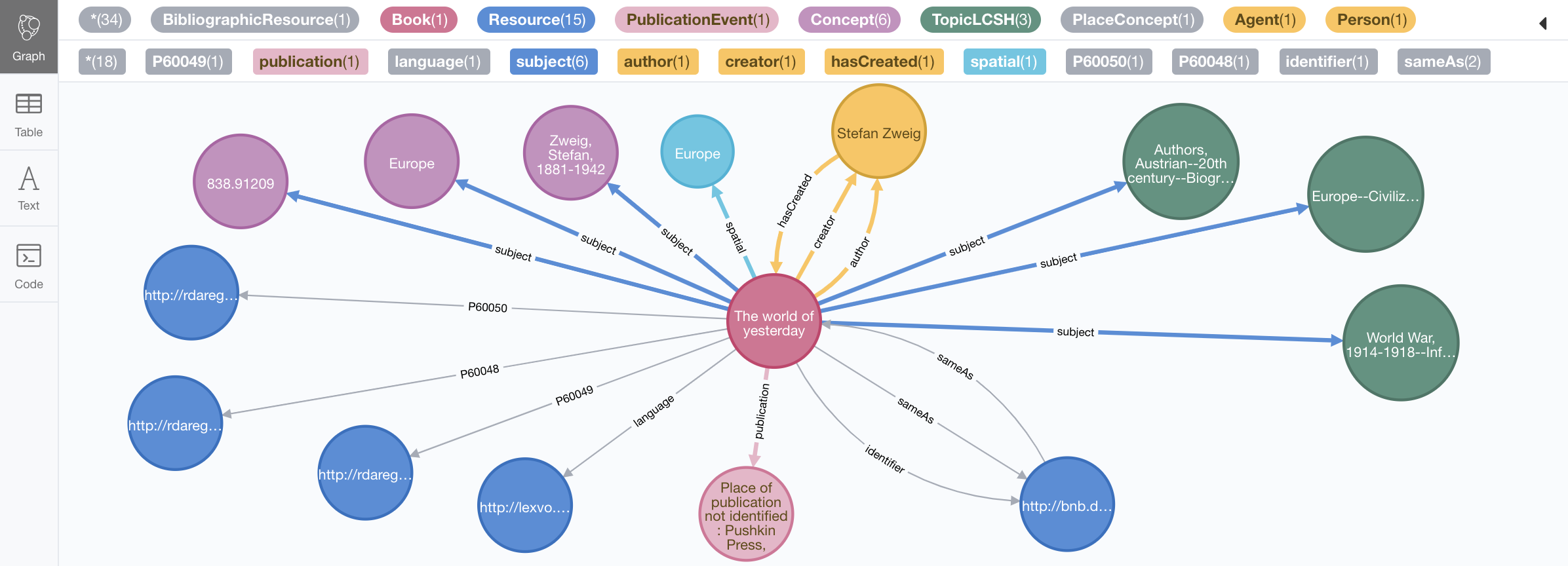
Of course you could do achieve this -or something similar- in different ways, in this case we are using a SPARQL CONSTRUCT query in order to be able to limit the number of triples returned for each resource as some of them are pretty dense.
Defining custom prefixes for namespaces
When applying url shortening on RDF ingestion (either explicitly or implicitly), we have the option of letting neosemantics automatically assign prefixes to namespaces as they appear in the imported RDF. But before doing that, a few popular ones will be set with familiar prefixes. These include "http://www.w3.org/1999/02/22-rdf-syntax-ns#" prefixed as rdf and "http://www.w3.org/2004/02/skos/core#" prefixed as skos.
At any point you can check the prefixes in use by running the n10s.nsprefixes.list procedure.
call n10s.nsprefixes.list();Before running your first import this method should return no results but after your first run, it should return a list containing at least the following entries.
| prefix | namespace |
|---|---|
"skos" |
"http://www.w3.org/2004/02/skos/core#" |
"sch" |
"http://schema.org/" |
"sh" |
"http://www.w3.org/ns/shacl#" |
"rdfs" |
"http://www.w3.org/2000/01/rdf-schema#" |
"dc" |
"http://purl.org/dc/elements/1.1/" |
"dct" |
"http://purl.org/dc/terms/" |
"rdf" |
"http://www.w3.org/1999/02/22-rdf-syntax-ns#" |
"owl" |
"http://www.w3.org/2002/07/owl#" |
Let’s say the RDF dataset that you are going to import uses the namespace http://neo4j.org/voc/sw# and you want it to be prefixed as neo instead of ns0 (or ns7) as would happen if the prefix was assigned automatically by neosemantics.
You can do this by calling the addNamespacePrefix procedure as follows:
CALL n10s.nsprefixes.add("neo", "http://neo4j.org/vocab/sw#");| prefix | namespace |
|---|---|
"neo" |
"http://neo4j.org/vocab/sw#" |
And then when the namespace is detected during the ingestion of the RDF data, the neo prefix will be used.
Make sure you know what you’re doing if you manipulate the prefix definition, especially after loading RDF data as you can overwrite namespaces in use, which would affect the possibility of regenerating the imported RDF.
Sometimes we have an RDF header with a bunch of prefix definitions.
This can come from a SPARQL query, from a Turtle document header or from an RDF/XML document root.
You can grab this fragment from your RDF document and pass it to the (experimental) addNamespacePrefixesFromText method and it will try to extract the namespace definitions and add each of them individually by invoking n10s.nsprefixes.addFromText.
Here’s an example of how it works:
WITH '<rdf:RDF xmlns:rdf="http://www.w3.org/1999/02/22-rdf-syntax-ns#"
xmlns:foaf="http://xmlns.com/foaf/0.1/"
xmlns:cert="http://www.w3.org/ns/auth/cert#"
xmlns:dct="http://purl.org/dc/terms/"' as txt
CALL n10s.nsprefixes.addFromText(txt) yield prefix, namespace
RETURN prefix, namespace;