Inferencing/Reasoning
By inferencing/reasoning we understand the process of getting information from the Neo4j database that is not explicitly stored. Here is a simple example: you have in your Neo4j graph some nodes labeled as loans and some nodes labeled as mortgages. If you manage to express the fact that a mortgage is a type of loan and consequently nodes labeled as mortgages are loans too, then you could expect your smart Neo4j DB to apply this reasoning on the fly and return both loan and mortgage nodes when you query for loans (even though you never explicitly labeled mortgage nodes as loans).
This kind of reasoning/inferencing is what this set of procedures will help you with.
The examples in this section will help understand how the different procedures and functions for inferencing work but check the Reference section for a complete list of methods and configuration parameters.
Hierarchies of Categories
To model a hierarchy of categories we’ll typically use nodes in the graph to represent the categories and
related nodes connected through SUBCAT_OF or NARROWER_THAN relationships (or whatever your
choice of terminology will be).
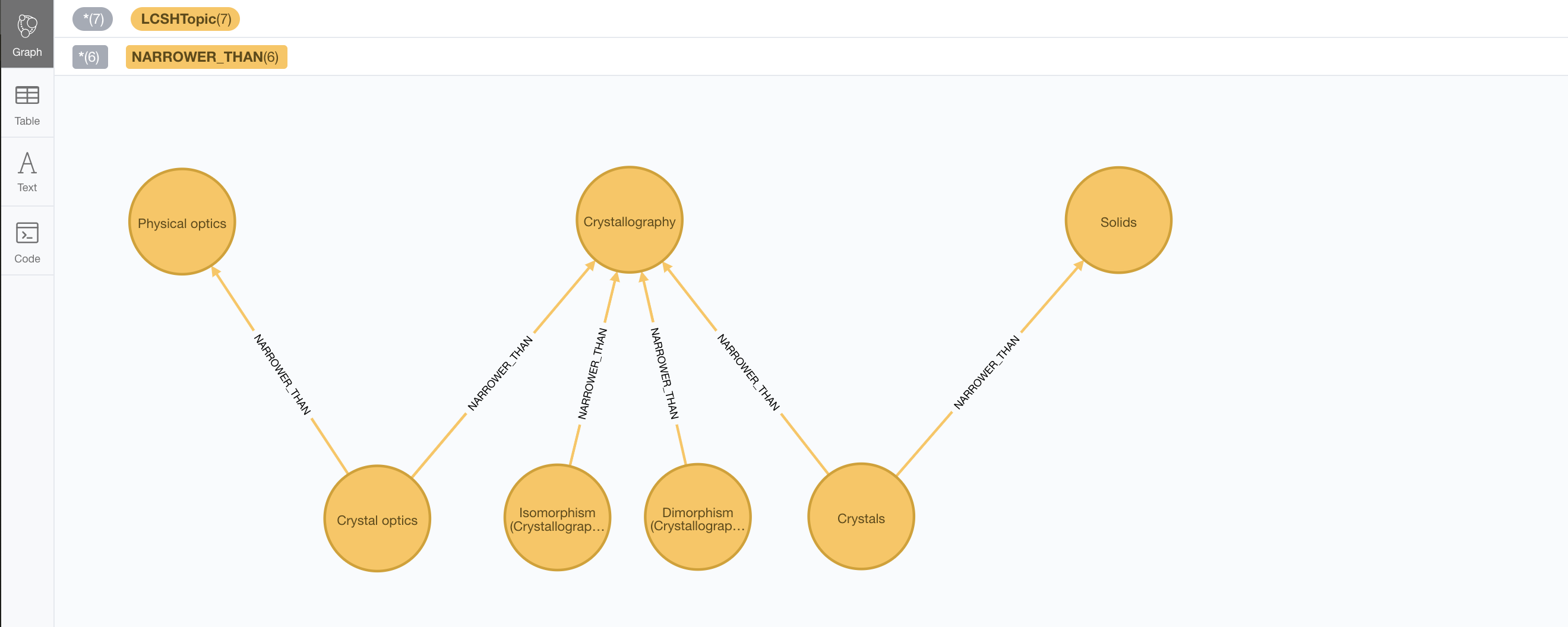
Here’s a set of categories from the Library of Congress Subject Headings.
CREATE (c:LCSHTopic { authoritativeLabel: "Crystallography", dbLabel: "Crystallography", identifier: "sh 85034498" })
CREATE (po:LCSHTopic { authoritativeLabel: "Physical optics", dbLabel: "PhysicalOptics", identifier: "sh 85095187" })
CREATE (s:LCSHTopic { authoritativeLabel: "Solids", dbLabel: "Solids", identifier: "sh 85124647" })
CREATE (c)<-[:NARROWER_THAN]-(:LCSHTopic { authoritativeLabel: "Crystal optics", dbLabel: "CrystalOptics", identifier: "sh 85034488" })-[:NARROWER_THAN]->(po)
CREATE (c)<-[:NARROWER_THAN]-(:LCSHTopic { authoritativeLabel: "Crystals", dbLabel: "Crystals", identifier: "sh 85034503" })-[:NARROWER_THAN]->(s)
CREATE (c)<-[:NARROWER_THAN]-(:LCSHTopic { authoritativeLabel: "Dimorphism (Crystallography)", dbLabel: "DimorphismCrystallography", identifier: "sh 2007001101" })
CREATE (c)<-[:NARROWER_THAN]-(:LCSHTopic { authoritativeLabel: "Isomorphism (Crystallography)", dbLabel: "IsomorphismCrystallography", identifier: "sh 85068653" })In this example we use LCSHTopic to label the categories and the NARROWER_THAN to link them in a
hierarchy that as we can see in this fragment does not necessarily need to be a tree (it the general
case it will be a graph).
We have defined a hierarchy of categories and now we’ll want to annotate individuals with the categories defined. To do this we have two main options:
-
We can use labels to tag a node representing an individual with the category it belongs to. While this approach is preferable in many cases, it will be harder to navigate to nodes with related labels (by related in this case I mean super or sublabels).
-
We can link nodes representing individuals to the category (or categories) they belong to using a
TYPEorIN_CATEGORY(or again whatever your preferred name for that relationship).
The following methods will help you leveraging explicit class hierarchies in your graph to run inferences whatever the modeling approach you follow from the ones described before.
n10s.inference.nodesLabelled
Let’s look at the first way of annotating individuals. This script creates a few publications (books) from the British National Library catalog sets a label on each of them. The labels match categories defined before.
CREATE (:Book:CrystalOptics { title: "Crystals and light", identifier: "2167673"})
CREATE (:Book:CrystalOptics { title: "Optical crystallography", identifier: "11916857"})
CREATE (:Book:IsomorphismCrystallography { title: "Isomorphism in minerals", identifier: "8096161"})
CREATE (:Book:Crystals { title: "Crystals and life", identifier: "12873809"})
CREATE (:Book:Crystals { title: "Highlights in applied mineralogy", identifier: "20234576"})Note that in this case there is no relationship connecting the books with the category they belong to. We are using labels instead. But there is an explicit hierarchy for these labels that we want to exploit.
What we want now is to be able to ask Neo4j for all the books on Crystallography and get all those
actually labelled as Crystallography but also all those labelled as any of Crystallography’s subcategories.
That’s exactly what the n10s.inference.nodesLabelled does for us. All we need to do is pass
the details on how is the category hierarchy built: catLabel will contain the label used to describe
categories (the default is Label) which in our case is LCSHTopic. We’ll also need to specify in the
subCatRel parameter, the relationship used to define the hierarchy (the default is SLO for
Sub Label Of) which in our case is NARROWER_THAN. Finally, we need to specify the name in the
category node containing the label name (the default is name) which in our example is dbLabel.
CALL n10s.inference.nodesLabelled('Crystallography', {
catNameProp: "dbLabel",
catLabel: "LCSHTopic",
subCatRel: "NARROWER_THAN"
})
YIELD node
RETURN node.identifier as id, node.title as title, labels(node) as categories;When we run this query, and even thoug not a single node in our graph is actually labelled as Crystallography,
we get the following results:
| id | title | categories |
|---|---|---|
"2167673" |
"Crystals and light" |
["CrystalOptics","Book"] |
"11916857" |
"Optical crystallography" |
["CrystalOptics","Book"] |
"12873809" |
"Crystals and life" |
["Crystals","Book"] |
"20234576" |
"Highlights in applied mineralogy" |
["Crystals","Book"] |
"8096161" |
"Isomorphism in minerals" |
["IsomorphismCrystallography","Book"] |
n10s.inference.hasLabel
If what we are looking for is not an set of nodes in a given category but rather a predicate telling us
whether a node is or not is in a category then the function we need is n10s.inference.hasLabel.
Let’s create a user with interests in some of the books in our database:
MERGE (jb:User { userId : "JB2020"}) with jb
MATCH (book1:Book { identifier : "20234576" })
MATCH (book2:Book { identifier : "11916857" })
WITH jb, book1, book2
CREATE (book1)<-[:INTERESTED_IN]-(jb)-[:INTERESTED_IN]->(book2)Now we can query for the books about Physical optics that he’s interested in.
Let’s first setup a parameter that defines the inference query:
:param inferenceParams => ({ catNameProp: "dbLabel", catLabel: "LCSHTopic", subCatRel: "NARROWER_THAN" });And now we can run our query:
MATCH (:User { userId : "JB2020"})-[:INTERESTED_IN]->(b:Book)
WHERE n10s.inference.hasLabel(b,'PhysicalOptics',$inferenceParams)
RETURN b.identifier as id, b.title as title, labels(b) as categories;And again, even though there’s no book explicitly labelled as 'PhysicalOptics', the query will produce the following result:
| id | title | categories |
|---|---|---|
"11916857" |
"Optical crystallography" |
["Book", "CrystalOptics"] |
Check the Reference section for a complete list of methods and configuration parameters for these methods and functions.
n10s.inference.nodesInCategory
Now let’s look at the second way of annotating individuals.
| If you were running the previous example delete all Book nodes before continuing with this second approach. |
This script creates a few of them and links them to the categories defined before.
MATCH (co:LCSHTopic { authoritativeLabel: "Crystal optics"})
MATCH (is:LCSHTopic { authoritativeLabel: "Isomorphism (Crystallography)"})
MATCH (cr:LCSHTopic { authoritativeLabel: "Crystals"})
CREATE (:Work { title: "Crystals and light", identifier: "2167673"})-[:HAS_SUBJECT]->(co)
CREATE (:Work { title: "Optical crystallography", identifier: "11916857"})-[:HAS_SUBJECT]->(co)
CREATE (:Work { title: "Isomorphism in minerals", identifier: "8096161"})-[:HAS_SUBJECT]->(is)
CREATE (:Work { title: "Crystals and life", identifier: "12873809"})-[:HAS_SUBJECT]->(cr)
CREATE (:Work { title: "Highlights in applied mineralogy", identifier: "20234576"})-[:HAS_SUBJECT]->(cr);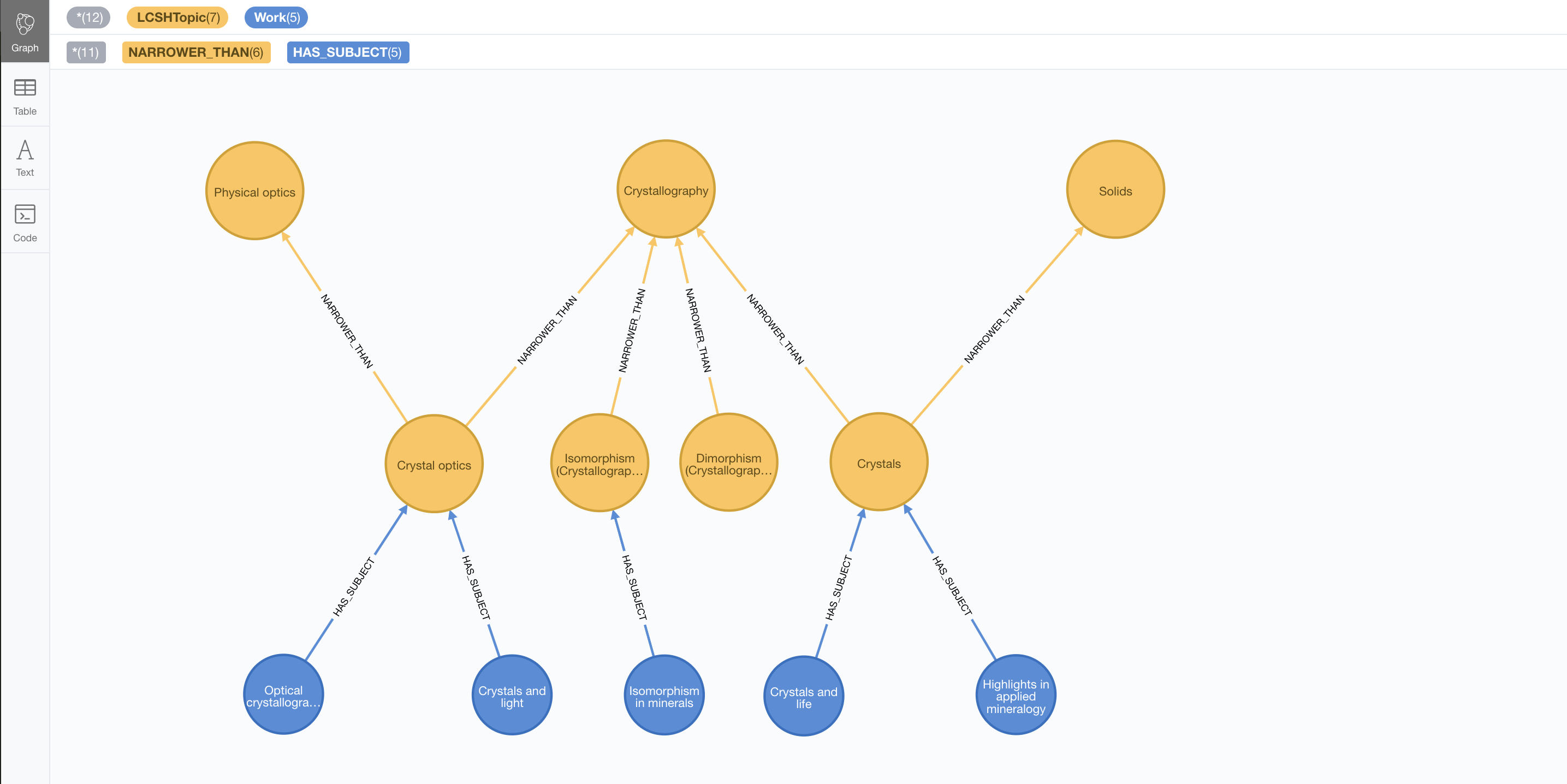
In this case, the query to get the nodes in a particular category will make use of
the n10s.inference.nodesInCategory procedure. This procedure takes as
parameters, the details of how is the category hierarchy built and how are individuals connected to
the categories: inCatRel specifies the relationship used to link an instance to a category (the
default is IN_CAT) which in our example is HAS_SUBJECT. subCatRel specifies the relationship used
to define the hierarchy (the default is SCO for Sub Category Of) which in our example is NARROWER_THAN.
MATCH (cat:LCSHTopic { authoritativeLabel: "Crystallography"})
CALL n10s.inference.nodesInCategory(cat, { inCatRel: "HAS_SUBJECT", subCatRel: "NARROWER_THAN"}) yield node
return node.title as work;When we run this Cypher fragment, we get the following list of results, even though not a single node in the graph is actually explicitly connected to the Crystallography category.
| work |
|---|
"Isomorphism in minerals" |
"Crystals and life" |
"Highlights in applied mineralogy" |
"Optical crystallography" |
"Crystals and light" |
n10s.inference.inCategory(node, category, {})
If what we are looking for is not an set of nodes in a given category but rather a predicate telling us
whether a node is or not is in a category then the function we need is n10s.inference.inCategory.
Let’s create a user with interests in some of the books in our database:
MERGE (jb:User { userId : "JB2020"}) with jb
MATCH (book1:Work { identifier : "20234576" })
MATCH (book2:Work { identifier : "11916857" })
WITH jb, book1, book2
CREATE (book1)<-[:INTERESTED_IN]-(jb)-[:INTERESTED_IN]->(book2);Now we can query for the books about Physical optics that he’s interested in.
Let’s update our inference parameter by running the following query:
:param inferenceParams => ({ inCatRel: "HAS_SUBJECT", subCatRel: "NARROWER_THAN"});And now we can run the query:
MATCH (phyOpt:LCSHTopic { authoritativeLabel: "Physical optics"})
MATCH (:User { userId : "JB2020"})-[:INTERESTED_IN]->(b:Work)
WHERE n10s.inference.inCategory(b,phyOpt,$inferenceParams)
RETURN b.identifier as id, b.title as title;And again, even though there’s no book explicitly connected to the 'PhysicalOptics' category, the query will produce the following result:
| id | title |
|---|---|
"11916857" |
"Optical crystallography" |
Remember to check the Reference section for a complete list of methods and configuration parameters for these methods and functions.
A real world example
We can use the n10s.onto.import.fetch procedure to import the NCBI Taxon ontology.
This is an ontology representation of the National Center for Biotechnology Information (NCBI) organismal taxonomy.
It contains 1.8 million classes (Class) and 3.6 million subClass of (SCO) relationships.
CALL n10s.onto.import.fetch("http://purl.obolibrary.org/obo/ncbitaxon.owl","RDF/XML");| terminationStatus | triplesLoaded | triplesParsed | namespaces | extraInfo | configSummary |
|---|---|---|---|---|---|
"OK" |
5480841 |
12581469 |
null |
"" |
{} |
Let’s add to the hierarchy a few individuals. Some dogs (NCBITaxon_9615, "Canis lupus familiaris"):
CREATE (p:Person { name: "Mr. Doglover"}) WITH p
UNWIND [ { name: "Perdita" , dob: "30/11/2016"}, { name: "Toby" , dob: "14/03/2019"}, { name: "Lucky" , dob: "14/11/2018"}, { name: "Pongo" , dob: "4/10/2012"}] as doggy
CREATE (:Pet:NCBITaxon_9615 { name: doggy.name, dob: doggy.dob })-[:OWNER]->(p);And why not? some mice (NCBITaxon_10092, "Mus musculus domesticus"):
CREATE (p:Person { name: "Mr. Mouselover"}) WITH p
UNWIND [ { name: "Mickey" , dob: "30/11/2016"}, { name: "Minnie" , dob: "14/03/2019"}, { name: "Topo" , dob: "14/11/2018"}, { name: "Rastamouse" , dob: "4/10/2012"}] as mouse
CREATE (:Pet:NCBITaxon_10092 { name: mouse.name, dob: mouse.dob })-[:OWNER]->(p);If we’re looking for instances of mammals in our database, we’d look for nodes labelled as NCBITaxon_40674
("Mammalia"). Obviously no node has been labelled as mammal, but we expect Neosemantics to do the job for us.
CALL n10s.inference.nodesLabelled('NCBITaxon_40674',{ catLabel: "Class", subCatRel: "SCO" }) YIELD node
RETURN node.name as name, node.dob as dob, labels(node);Only a few milliseconds needed to identify them in the nearly 11k categories under Mammalia.
| name | dob | labels(node) |
|---|---|---|
"Mickey" |
"30/11/2016" |
["Pet","NCBITaxon_10092"] |
"Minnie" |
"14/03/2019" |
["Pet","NCBITaxon_10092"] |
"Topo" |
"14/11/2018" |
["Pet","NCBITaxon_10092"] |
"Rastamouse" |
"4/10/2012" |
["Pet","NCBITaxon_10092"] |
"Perdita" |
"30/11/2016" |
["NCBITaxon_9615","Pet"] |
"Toby" |
"14/03/2019" |
["NCBITaxon_9615","Pet"] |
"Lucky" |
"14/11/2018" |
["NCBITaxon_9615","Pet"] |
"Pongo" |
"4/10/2012" |
["NCBITaxon_9615","Pet"] |
Interestingly, and because Neo4j is a native Graph DB implementing index free adjacency, if we were
to search across the 1.2 million categories for all instances of "Eukaryota" (NCBITaxon_2759),
(one of the top three categories that all cellular organisms are divided into) it would take Neosemantics
exactly the same time to identify them. Here’s the query:
CALL n10s.inference.nodesLabelled('NCBITaxon_2759',{ catLabel: "Class", subCatRel: "SCO" }) YIELD node
RETURN node.name as name, node.dob as dob, labels(node)Similarly, we can verify in milliseconds how many of an individual’s pets are actually instances of "Eukaryota". Here’s how:
MATCH path = (:Person { name : "Mr. Doglover"})<-[:OWNER]-(pet)
WHERE n10s.inference.hasLabel(pet,'NCBITaxon_2759',$inferenceParams)
RETURN count(pet);Hierarchies of Relationships
Just like we did with categories, we can use rdfs:subPropertyOf to create hierarchies of relationships,
or in other words to state that all resources connected by one relationship are also implicitly connected
by any parent relationship. If We state that ACTED_IN is a subproperty of WORKED_IN,
when we find in the graph that Keanu Reeves ACTED_IN The Matrix, we can safely derive the fact that he
also WORKED_IN that movie, even if there is not an explicit WORKED_IN relationship in the graph
between Keanu and The Matrix.
This is useful in situations where we want to be able to dynamically define relationships by composing
existing ones.
The n10s.inference.getRels stored procedure uses exactly these semantics to infer implicit relationships between nodes in the graph.
n10s.inference.getRels
Let’s take the movie database.
Remember you can have it loaded in Neo4j by running :play movies and following the instructions in the guide.
Let’s say we have a fragment of a movie ontology that contains a definition of a relationship hierarchy.
It does it by defining a number of rdfs:subPropertyOf statements between relationships.
For instance, it states that every ACTED_IN relationship is also a WORKED_IN one.
This is the triple in question:
...
neovoc:ACTED_IN a owl:ObjectProperty;
rdfs:label "ACTED_IN";
rdfs:subPropertyOf neovoc:WORKED_IN .
...To see this inferencing procedure in action, we’ll start by loading the ontology.
We can do this by either using the n10s.onto.import.fetch or the n10s.rdf.import.fetch methods described in the Importing RDF Data section.
| We can get a hierarchy from an ontology or we can create it with a cypher script from any other source. |
If we run:
CALL n10s.onto.import.fetch("https://github.com/neo4j-labs/neosemantics/raw/3.5/docs/rdf/movieDBRelHierarchy.ttl", "Turtle");We should get a simple hierarchy of properties like the one in this screen capture from the Neo4j browser.
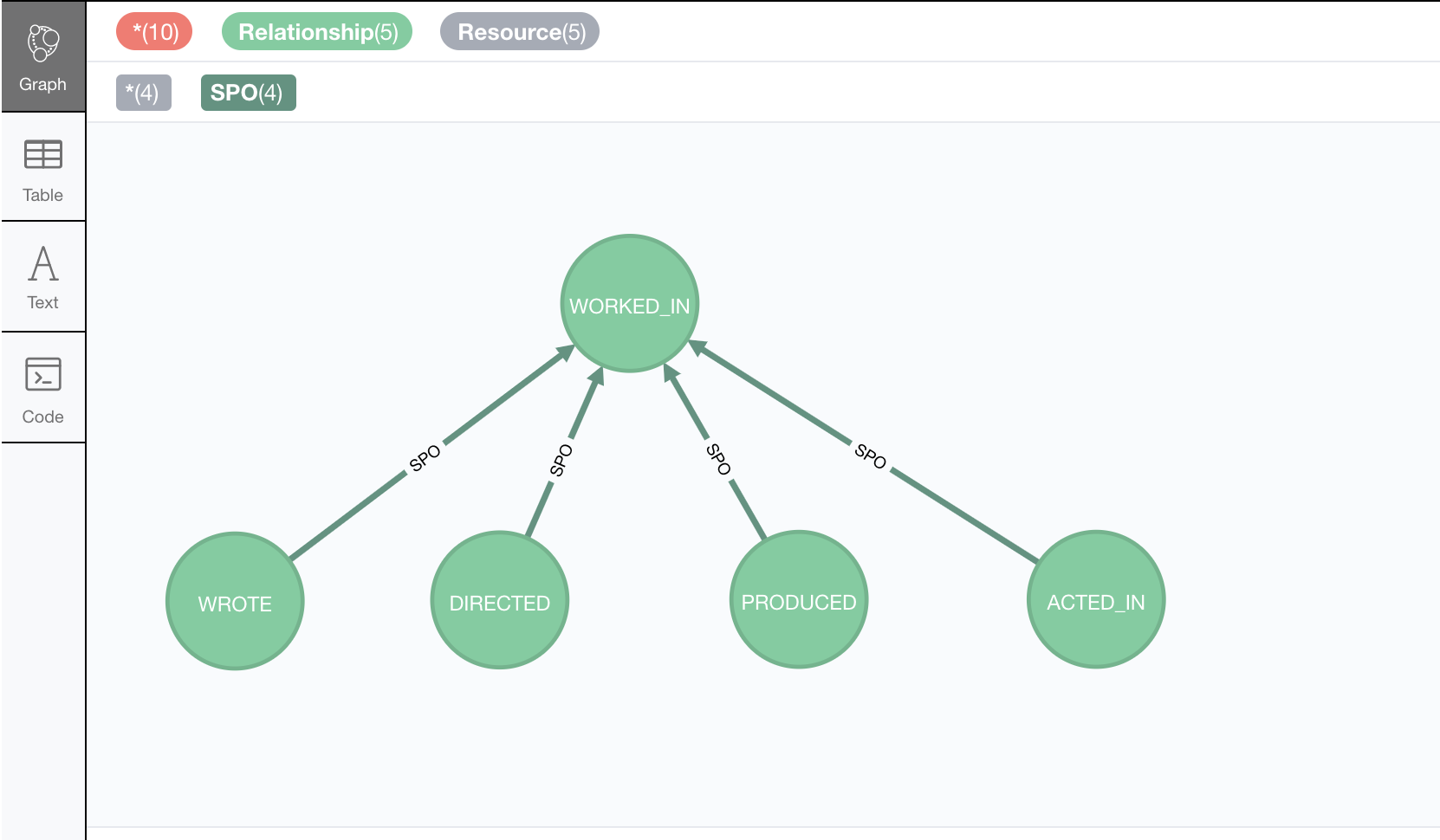
Writing a query that returns all nodes connected to the movie The Matrix through the 'virtual' WORKED_IN relationship
is an easy task with the n10s.inference.getRels procedure.
match (thematrix:Movie {title: "The Matrix"})
call n10s.inference.getRels(thematrix,"WORKED_IN", { subRelRel: "SPO" }) yield rel, node
return type(rel) as relType, node| relType | node |
|---|---|
"ACTED_IN" |
(:Person {name: "Keanu Reeves", born: 1964}) |
"PRODUCED" |
(:Person {name: "Joel Silver", born: 1952}) |
"DIRECTED" |
(:Person {name: "Lana Wachowski", born: 1965}) |
"ACTED_IN" |
(:Person {name: "Carrie-Anne Moss", born: 1967}) |
"ACTED_IN" |
(:Person {name: "Laurence Fishburne", born: 1961}) |
"ACTED_IN" |
(:Person {name: "Hugo Weaving", born: 1960}) |
"ACTED_IN" |
(:Person {name: "Emil Eifrem", born: 1978}) |
"DIRECTED" |
(:Person {name: "Lilly Wachowski", born: 1967}) |
Now let’s say we want to modify the meaning of the WORKED_IN relationship to exclude PRODUCED and
keep only artistic involvement connections, tis is WROTE, ACTED_IN and DIRECTED. We don’t need
to alter our database, just our ontology.
MATCH (:Relationship {name:"PRODUCED"})-[r:SPO]->(:Relationship {name:"WORKED_IN"})
DELETE rIf we run the same query again, we’ll get different results, this time excluding producers. Think of this in a large scale DB. We can effectively modify relationships globally by adding or deleting a simple link to the hierarchy and without having to modify every single instance.