Combining 3 Biochemical Datasets in a Graph Database

Back End Developer at CytoSMART
6 min read

The open measurement graph will be used to find connections between different measurements in different experiments and conditions. In a previous blog, we added chemical compounds — and their synonyms — from PubChem into the graph. In this blog, we are going to populate the database with NCI60 measurements.
Before we do that, we will first design the graph structure. Biomedical research is done in experiments, each holding multiple conditions. An experiment is a set-up, and a condition is a single instance of this set-up with one set of variables.
In the case of NCI60, the experiment is a cell growth medium that is measured for confluence at timepoint zero and 48 hours. The cell line and compound will be variable. A condition would be cell line A498 with compound NSC 19893.
In the case of NCI60, it has 5065 experiments. All of them are done as identically to the others as possible. However, slight variations can occur.
This includes:
– who did the experiment
– what were the values of everything NOT measured
– for biomedical research: how did the cell lines evolve between experiments?
This is why we note down the experiment.
Graph design
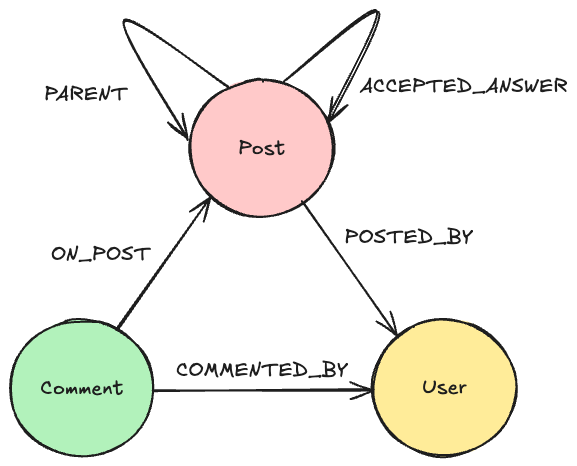
The PubChem graph has two node types and one relationship type.
(Synonym)-[:IS_ATTRIBUTE_OF]->(Compound)
We keep this in mind while designing, so the rest will follow a similar pattern.
An experiment has a few things it is always using (USES) — these are the constants of the experiment. An example of this is the protocol or the growth medium of the cells (compound). Connecting these to the experiment makes it clear they do not vary between conditions and prevent repetition. Some User OWNS this experiment.
The protocol has a synonym in between so different synonyms of the same protocol can be used, given that people rarely agree on a single name.
Next up are the Conditions, which are attributes of an experiment (IS_ATTRIBUTE_OF). This way, they have the same relationship as synonyms to compounds. A condition USES its variable’s synonyms (compound and cellLine from NCI60) and MEASURES a Measurement (like GI50). A condition is not limited to how many measurements it does, or to how many variables it uses.
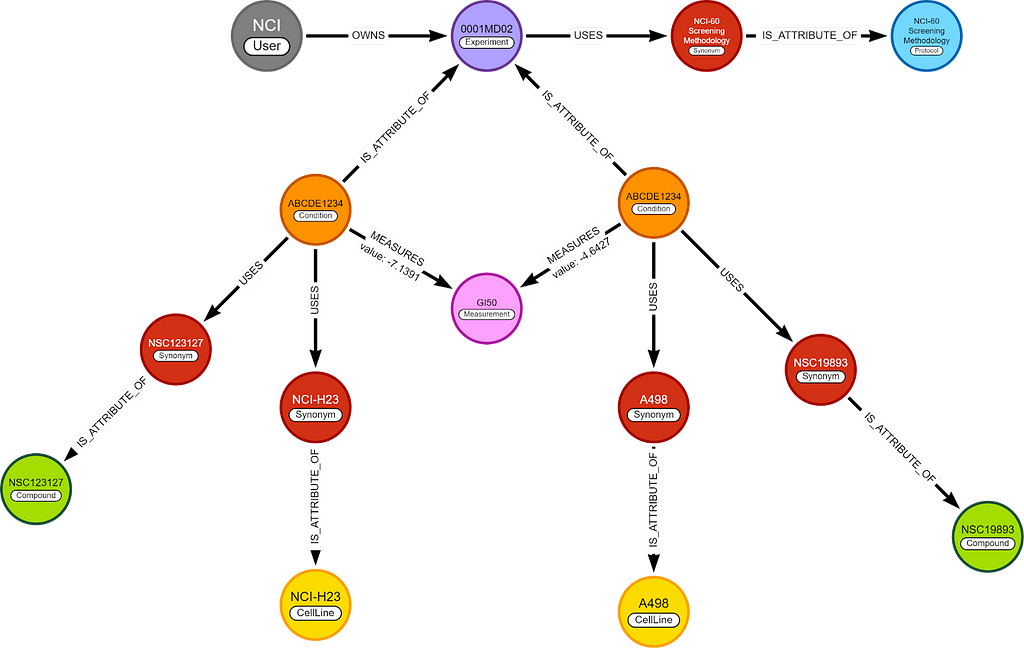
NCI60 dataset
NCI (National Cancer Institute, USA) has the NCI60 dataset. This dataset contains 50k+ chemicals tested on ~60 cell lines. Almost all of these cell lines are human cancer cell lines.
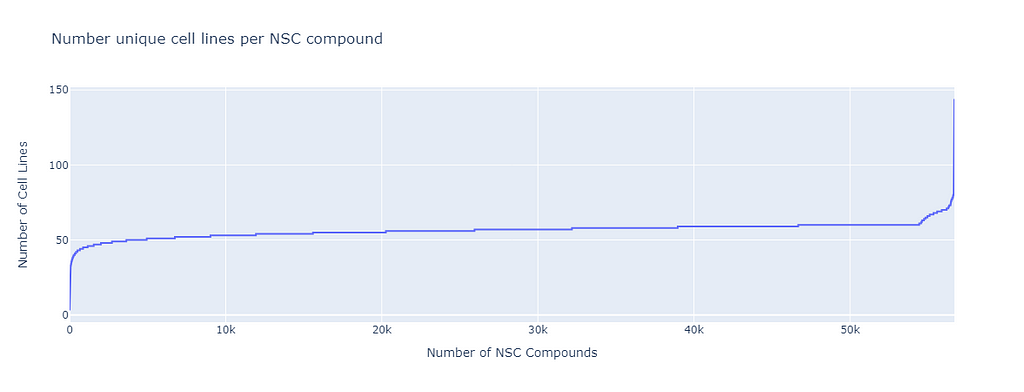
If we plot the NSC compounds against the number of unique Cell lines tested with that NSC compound, we find that most compounds are tested with at least 50 cell lines. This is for the GI50 part of the data (concentration of the compound needed for 50% Growth Inhibitor).
The missing cell lines can come from failed experiments since compounds are not fully tested if the effect is too small in the first “1 dose step’” of the protocol.
In order to add the experiment to the database, we need to do two things: figure out which synonym fits the NSC number, and introduce cell lines to connect to.
Introducing cell lines
Cell biologists like to make use of cell lines in their experiments. These are made, selected, or designed to last for many generations. The cell lines give consistency to the different experiments. These cell lines are also shared around the world, bringing different experiments closer to each other.
If we zoom in on cancer, we will find that no two cancers are the same. Every patient has unique cells, followed by a unique mutation that turned the cell into cancer. If researchers use these in their experiments, no one can repeat their findings unless they get the exact same cells. This is something that is not always possible because many cancer cells will die after a number of divisions. Other times, the patient did not give permission to share with other researchers.
Cancer cell lines are often immortal. A549 is one of these immortal lung cancer cell lines. If multiple experiments were done on A549, those could be more easily compared to each other than when every experiment used its own lung cancer cells.
The NCI60 experiments are also done on these kinds of cell lines. For the ontology, the Chembl cells RDF is used and loaded in with n10s. This gives us 2000 cell lines, 1466 are human cell lines.
While 2000 is very limited, the Chembl cells are easy to load in and can be connected to the Cellosaurus if I want to extend the cell lines.
Chembl taxonomy goes from the cell line directly to the species and it is not very useful for NCI60, given that those are almost all done on human cell lines — except for four conditions that use the famous CHO cells (Chinese hamster ovary cells).
With the help of a script, I selected the Chembl cell lines with the best match for the NCI60 cell lines. After that, a manual step follows to select the best match. The results are stored in a JSON.
Picking the NSC Synonym
In a previous blog, I connected and found all NSC numbers related to a compound. In there, we found an NSC synonym for 55k NSC numbers (e.q. “NSC 123” or “NSC-123” as synonyms for NSC number 123). However, connecting an NSC number to multiple synonyms could cause problems, because it seems like multiple chemicals are used.
The goal is to connect every NSC number to one — and only one — synonym.
Single compound
90.4% of the NSC numbers have only one compound shared between all its synonyms. For these, we pick the first synonym. This deals with 50k of 55k NSC numbers.
Update synonyms
In a previous blog, we found that not all synonyms are up-to-date. We are going to reuse that function again on the 9.6% that does not have a single compound. If we do NOT do this, the next part will only match 14 of 5287 NSC numbers.
Better connected one
In some cases, one synonym is connected to all compounds of all other synonyms. If this is the case, we pick this one. This also works if all synonyms are connected to multiple compounds. This increases our coverage to 97.9%.
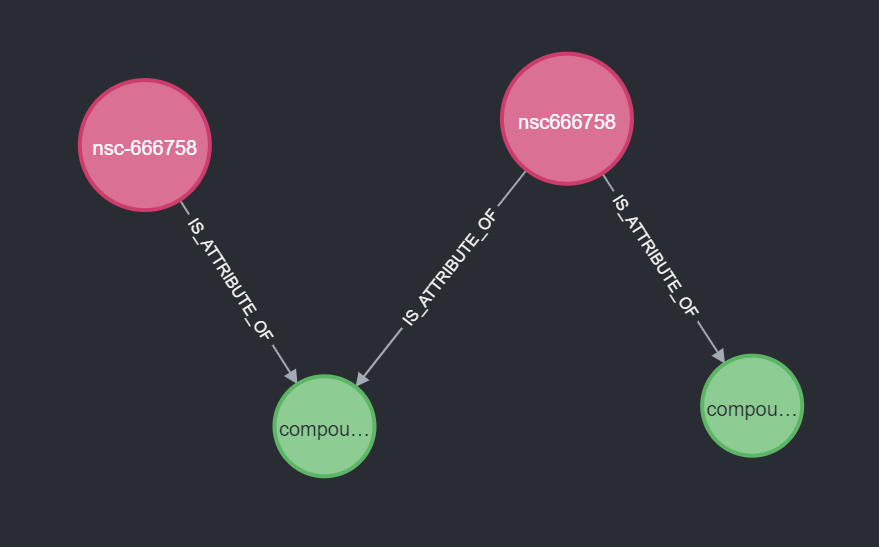
Another well-connected synonym
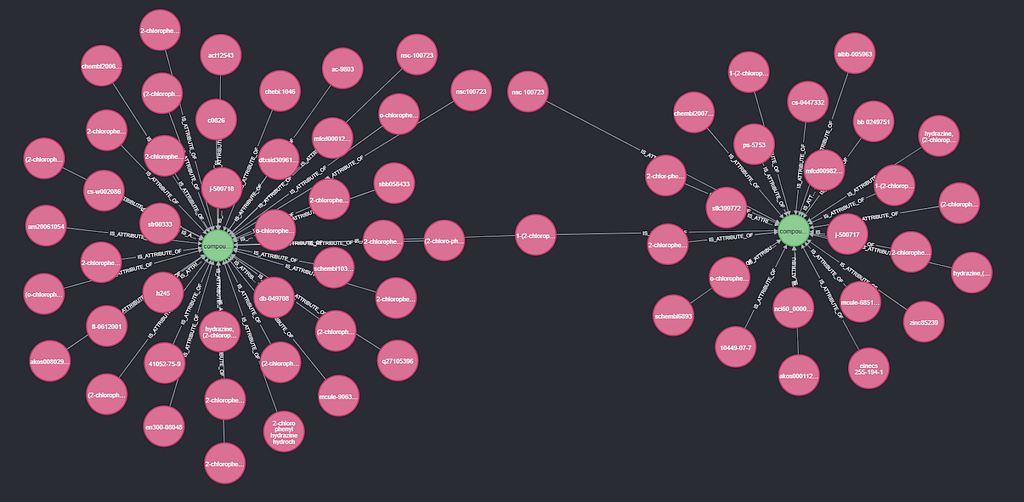
For the next 1144 nsc numbers, we still do not have a synonym picked. In this case, we are going to look for a synonym that connects all compounds, but this synonym does not have to be an NSC synonym.
Below, we see an example of nsc number 100723. The synonyms “nsc100723” and “nsc-100723” are connected to compound 1, but “nsc 100723” is connected to compound 2. None of the nsc synonyms connect to both compounds, but the synonym “1-(2-chlorophenyl) hydrazine hydrochloride” does. This matches another 503 synonyms, bringing the coverage to 98.8%.
The query is:
// Get all NSC synonyms
CALL {
CALL db.index.fulltext.queryNodes(‘synonymsFullText’, $nsc_query)
YIELD node, score
return node limit 10
}
// Get all compounds connect to the synonyms
MATCH (node)-[:IS_ATTRIBUTE_OF]->(c:Compound)
WITH collect(DISTINCT c) as compounds
// Find all synonyms connected to ALL compounds
UNWIND compounds as c
MATCH (s:Synonym)-[:IS_ATTRIBUTE_OF]->(c:Compound)
WHERE ALL(compound IN compounds WHERE (s)-[:IS_ATTRIBUTE_OF]->(compound))
// Return those synonyms
WITH DISTINCT s as s
RETURN s.name as name, s.pubChemSynId as synonymId
The last 641 NSC numbers
Let’s have a look at nsc 5038, one of the not matched NSC numbers. Its synonyms are connected to compounds 720071 and 221197, but these compounds are really not the same. Picking one or the other requires knowledge I do not have. This is the case for all random numbers I investigated. So this last 1.2% will be ignored for now.
Result
Using the filtering and selection above, the final script combines all three datasets. This results in 4m conditions, which are part of 4.6k experiments. In my next installment, I will explore the dataset.
Combining 3 Biochemical Datasets in a Graph Database was originally published in Neo4j Developer Blog on Medium, where people are continuing the conversation by highlighting and responding to this story.